EAM User Guide: Voter's Handbook Chapter
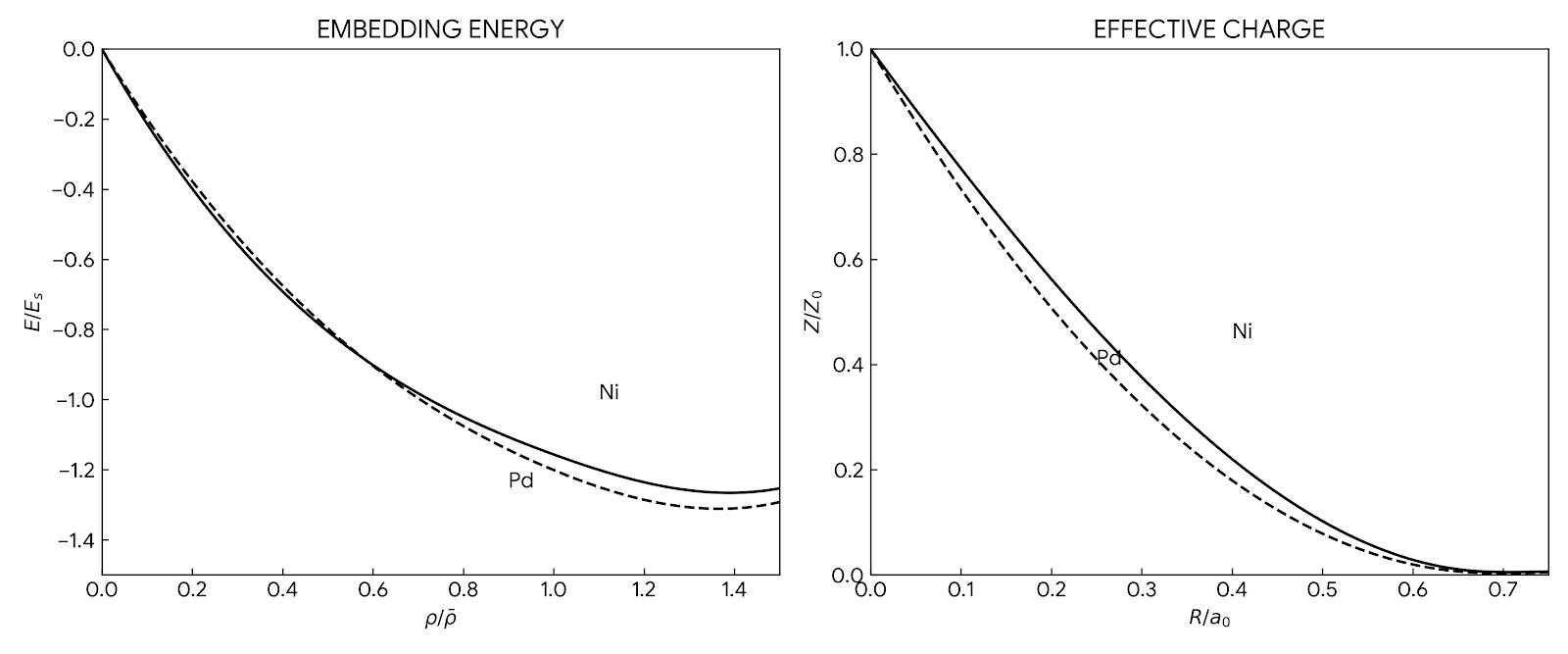
This seminal 1994 handbook chapter serves as a definitive user guide for the Embedded-Atom Method (EAM). It details the theoretical derivation from density-functional theory, synthesizes related methods like the Glue Model, and provides a complete tutorial on fitting potentials, illustrated with a specific implementation for the Ni-Al-B system.