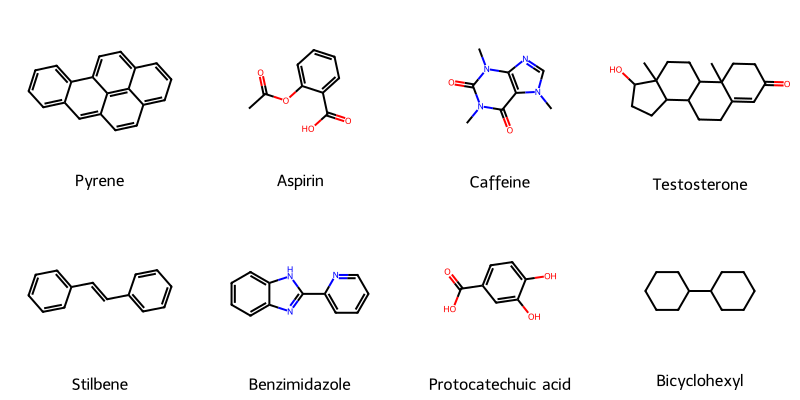
Molecular String Renderer: Robust Visualization Infrastructure
A robust, type-safe Python library for converting chemical string representations (SMILES, SELFIES, InChI) into …...

A robust, type-safe Python library for converting chemical string representations (SMILES, SELFIES, InChI) into …...
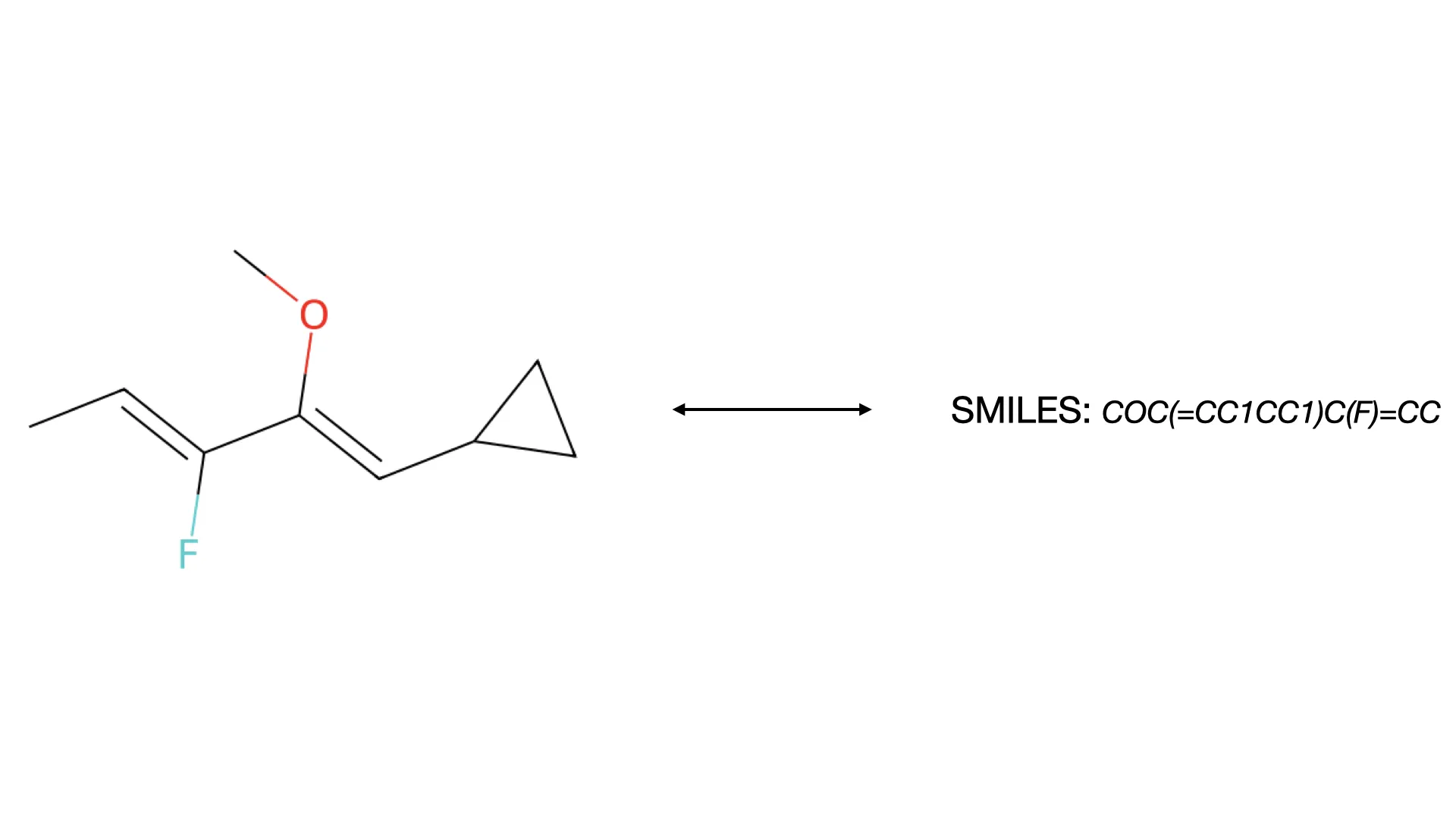
A micro-review of Optical Chemical Structure Recognition (OCSR), tracing its evolution from rule-based systems to …
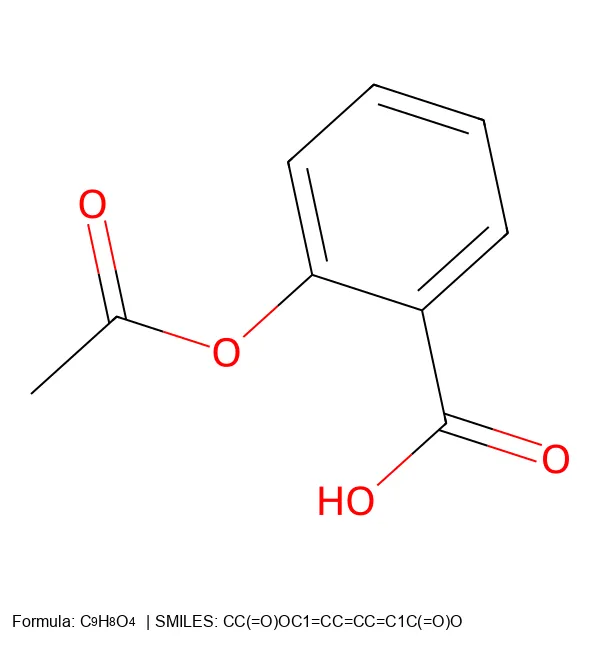
Learn how to create 2D molecular images from SMILES strings using RDKit and PIL, with proper formatting and legends.

The Müller-Brown potential: a classic two-dimensional analytical benchmark for testing optimization algorithms, reaction …...

Compare inverse transform sampling and von Neumann's rejection method for exponential random numbers with Python …

High-performance, GPU-accelerated PyTorch framework for the Müller-Brown potential, featuring JIT compilation, …...

Guide to implementing the Müller-Brown potential in PyTorch, comparing analytical vs automatic differentiation with …
Daw and Baskes's foundational 1984 paper introducing the Embedded-Atom Method (EAM), a many-body potential for metal …...
Torrie and Valleau's 1977 paper introducing Umbrella Sampling, an importance sampling technique for Monte Carlo …...
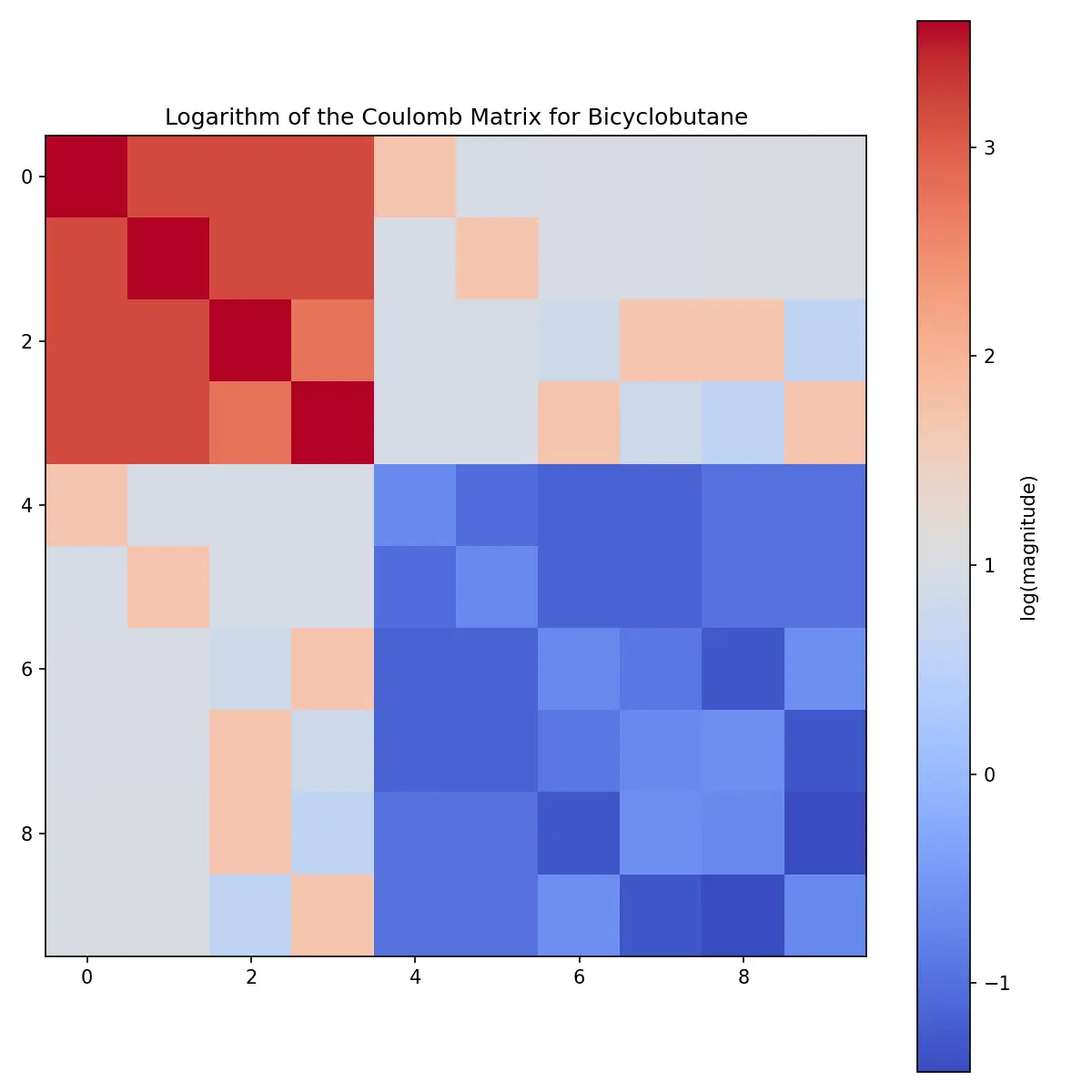
Learn how Coulomb matrices encode 3D molecular structure for machine learning from basic theory to Python implementation …
What happens to bills in Congress? Analyzing 15K+ bills from the 117th Congress to understand legislative patterns, …

Learn about the Kabsch algorithm for optimal point alignment with implementations in NumPy, PyTorch, TensorFlow, and JAX …