String Representations for Chemical Image Recognition
Ablation study comparing SMILES, DeepSMILES, SELFIES, and InChI for OCSR. SMILES achieves highest accuracy; SELFIES …
Ablation study comparing SMILES, DeepSMILES, SELFIES, and InChI for OCSR. SMILES achieves highest accuracy; SELFIES …
Deep learning model using Swin Transformer and Focal Loss for OCSR, achieving 98.58% accuracy on synthetic benchmarks.
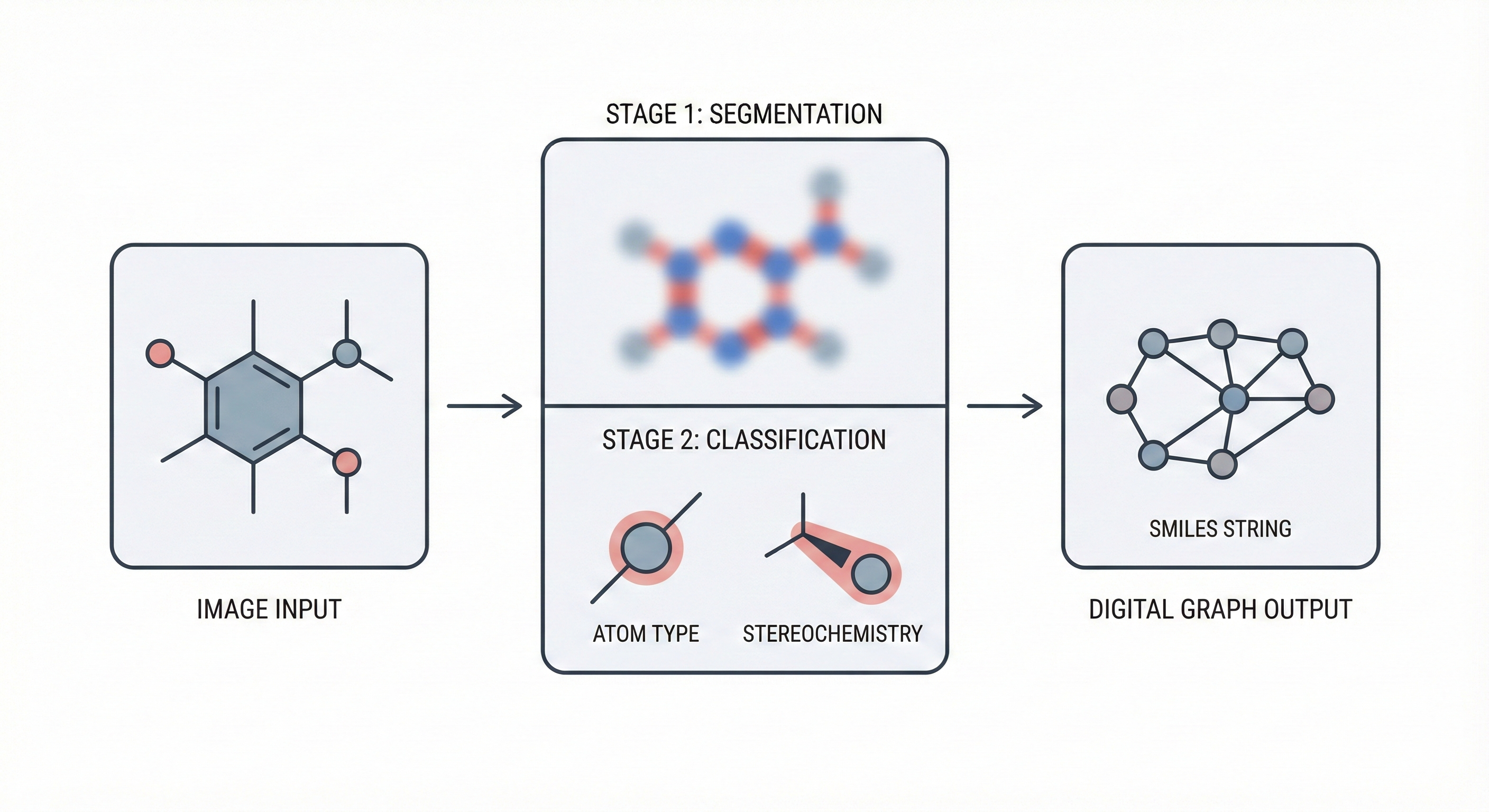
Deep learning OCSR method using semantic segmentation and classification CNNs to reconstruct chemical graphs with …
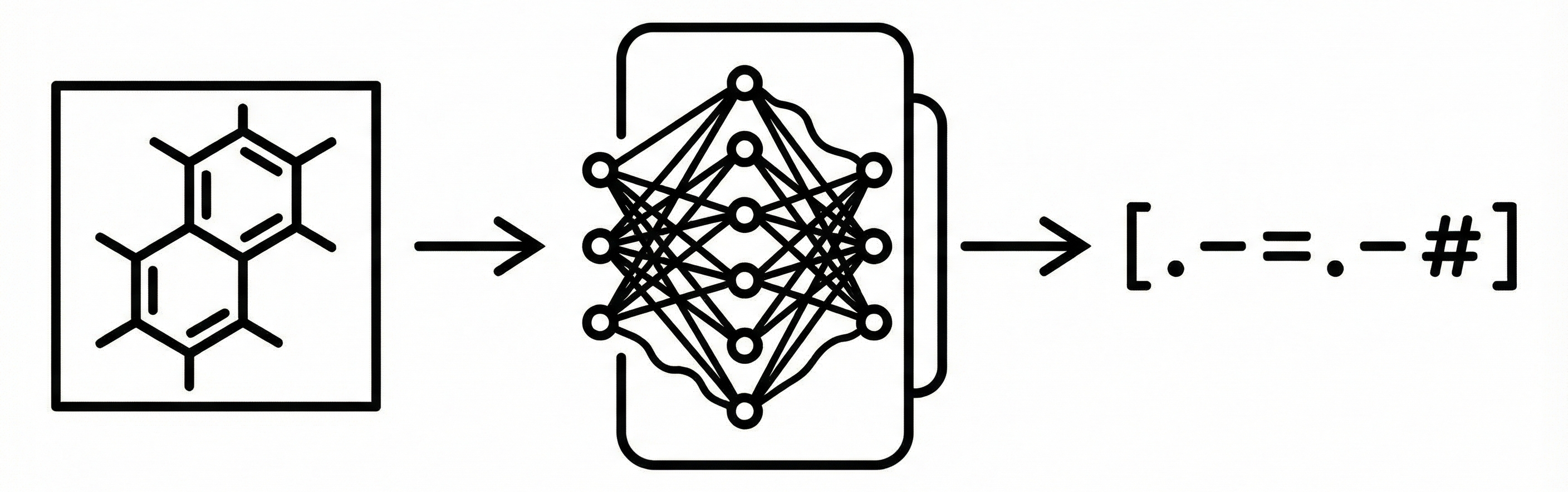
Deep learning method for optical chemical structure recognition using image captioning networks trained on millions of …
An end-to-end deep learning approach using U-Net and CNN-LSTM to segment and predict chemical structures from document …
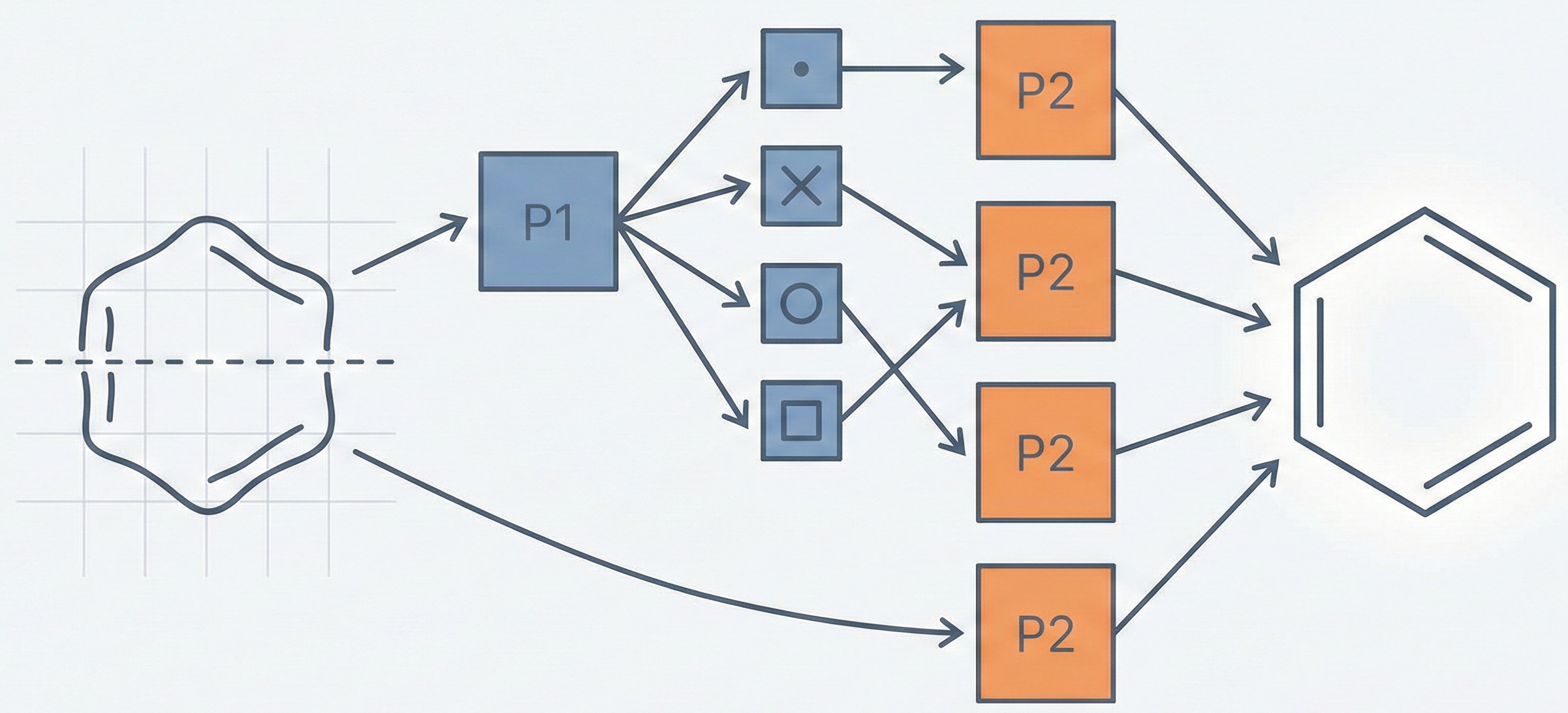
A two-phase neural network approach for recognizing handwritten heterocyclic chemical rings with ~94% accuracy.
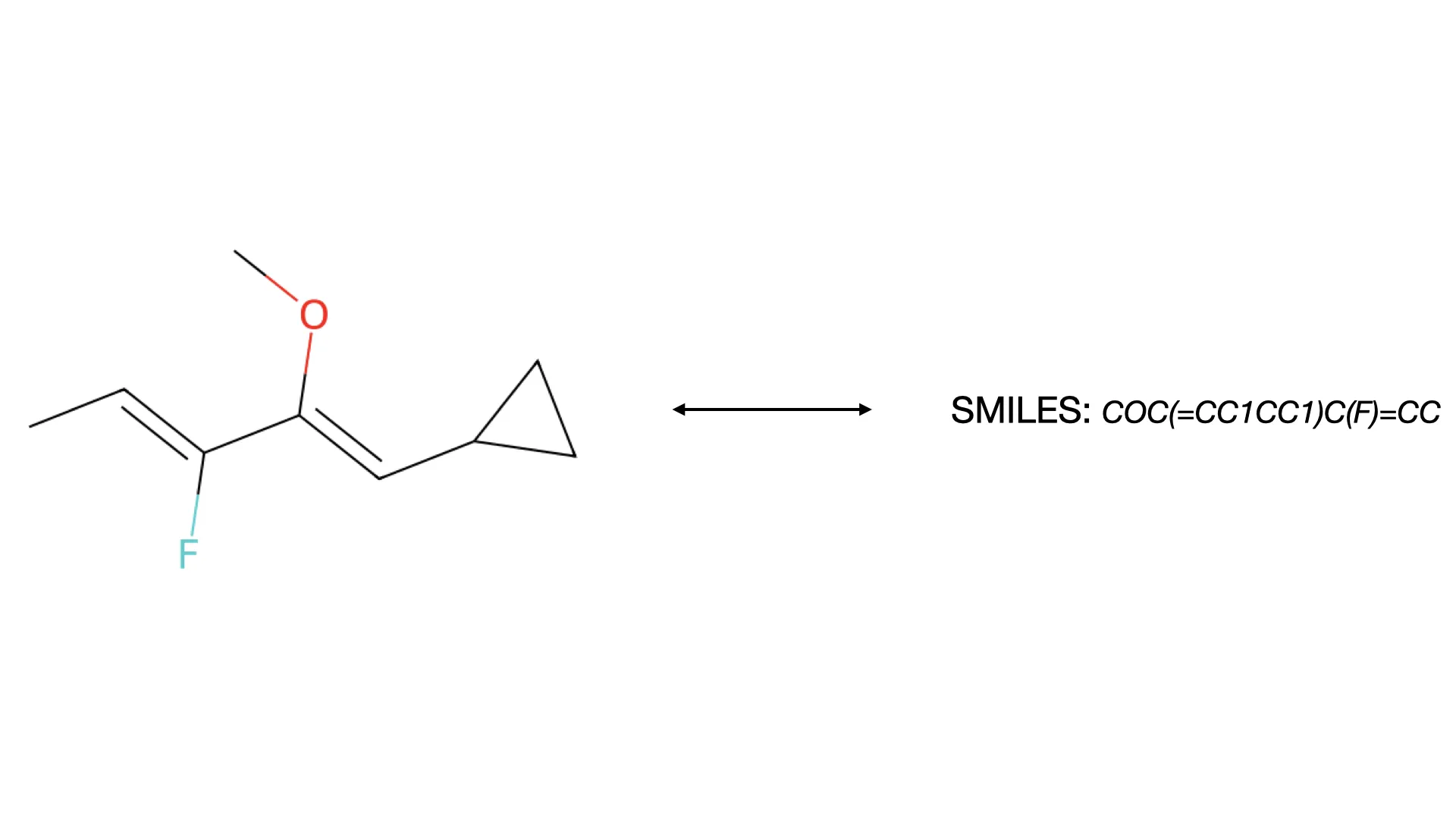
Two-stage CNN approach for converting molecular images to SMILES using CDDD embeddings and extensive data augmentation.
Foundational OCSR method combining neural OCR with chemical rule-based post-processing for automated structure …
Machine vision approach using Gabor wavelets and Kohonen networks to classify chemical raster images and extract …

Hinton's 1984 technical report establishing the theoretical efficiency of distributed representations over local …

Campos & Ji's method for converting 2D molecular images to SMILES strings using Transformers and SELFIES representation.
A seminal 1992 system for Optical Chemical Structure Recognition (OCSR) using neural networks and heuristic graph …