ChemDFM-X: Large Multimodal Model for Chemistry
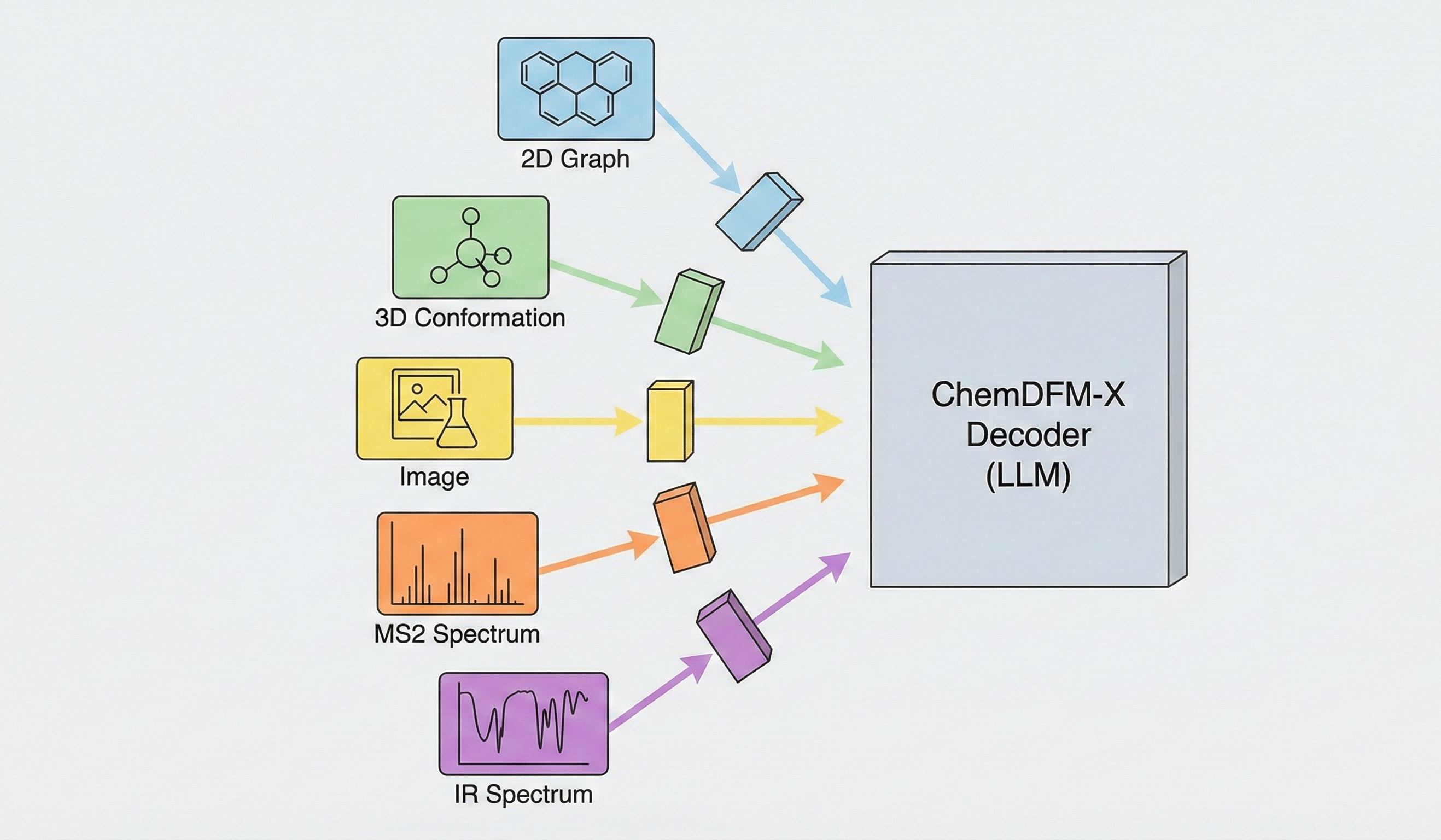
Multimodal chemical model integrating 5 modalities (2D graphs, 3D conformations, images, MS2/IR spectra) trained on 7.6M …

Multimodal chemical model integrating 5 modalities (2D graphs, 3D conformations, images, MS2/IR spectra) trained on 7.6M …
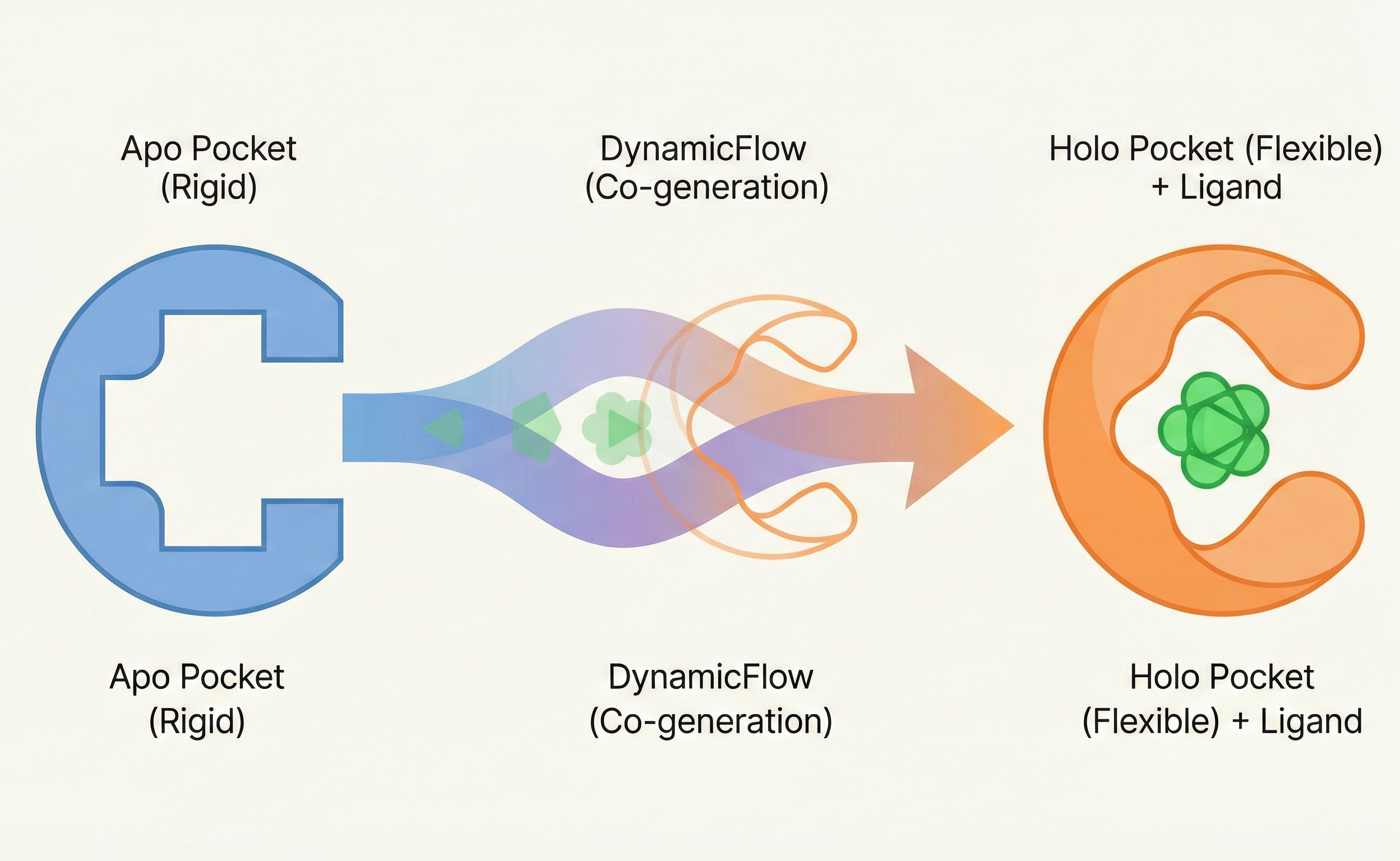
Flow matching model that co-generates ligands and flexible protein pockets, addressing rigid-receptor limitations in …
Comparative analysis of image-to-sequence OCSR methods across architecture, output format, training data, and compute …
A multi-modal LLM aligning 2D molecular graphs with text via two-stage instruction tuning for drug discovery tasks.
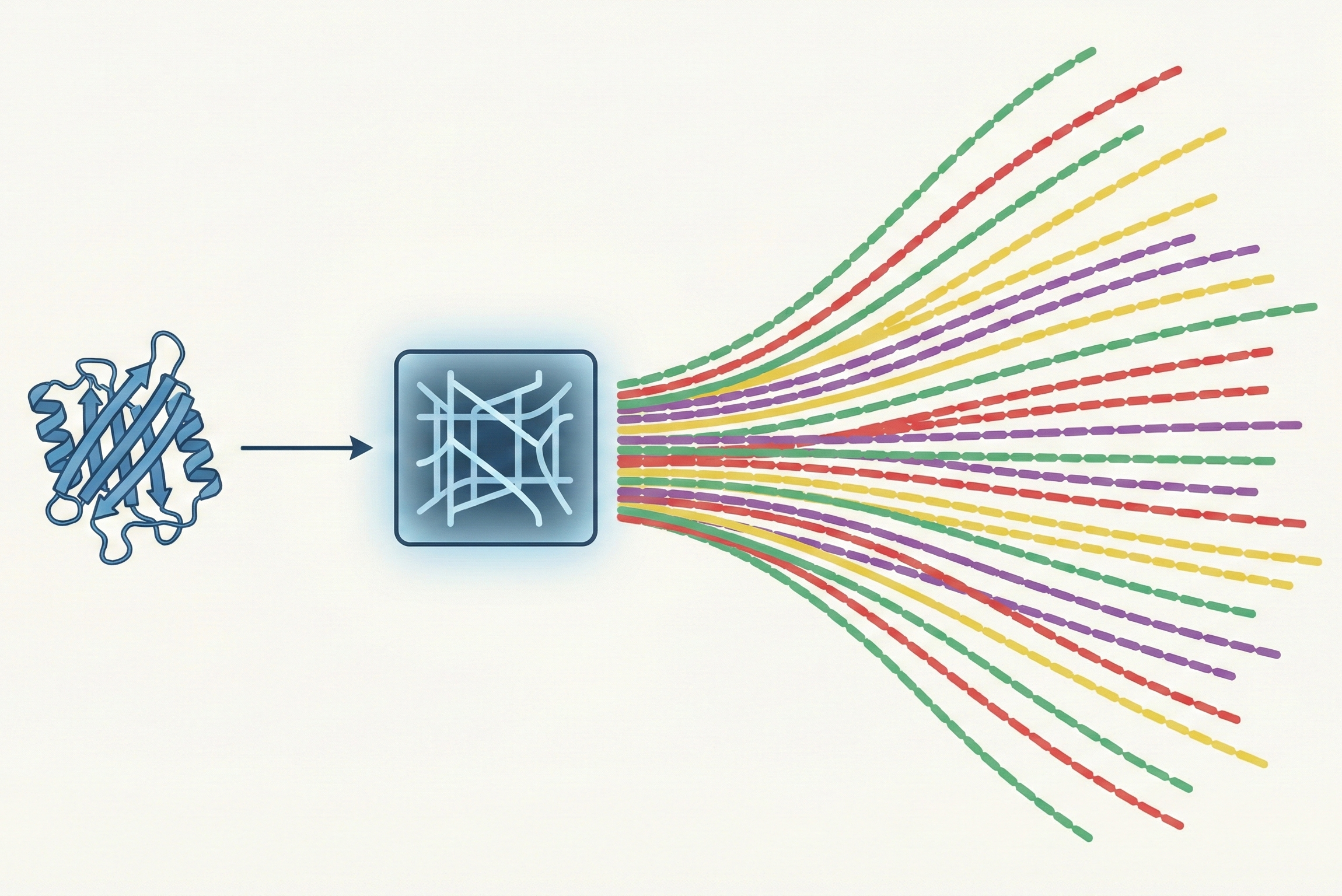
A fast, diverse inverse folding method combining deep learning with Potts models to capture full sequence landscapes.
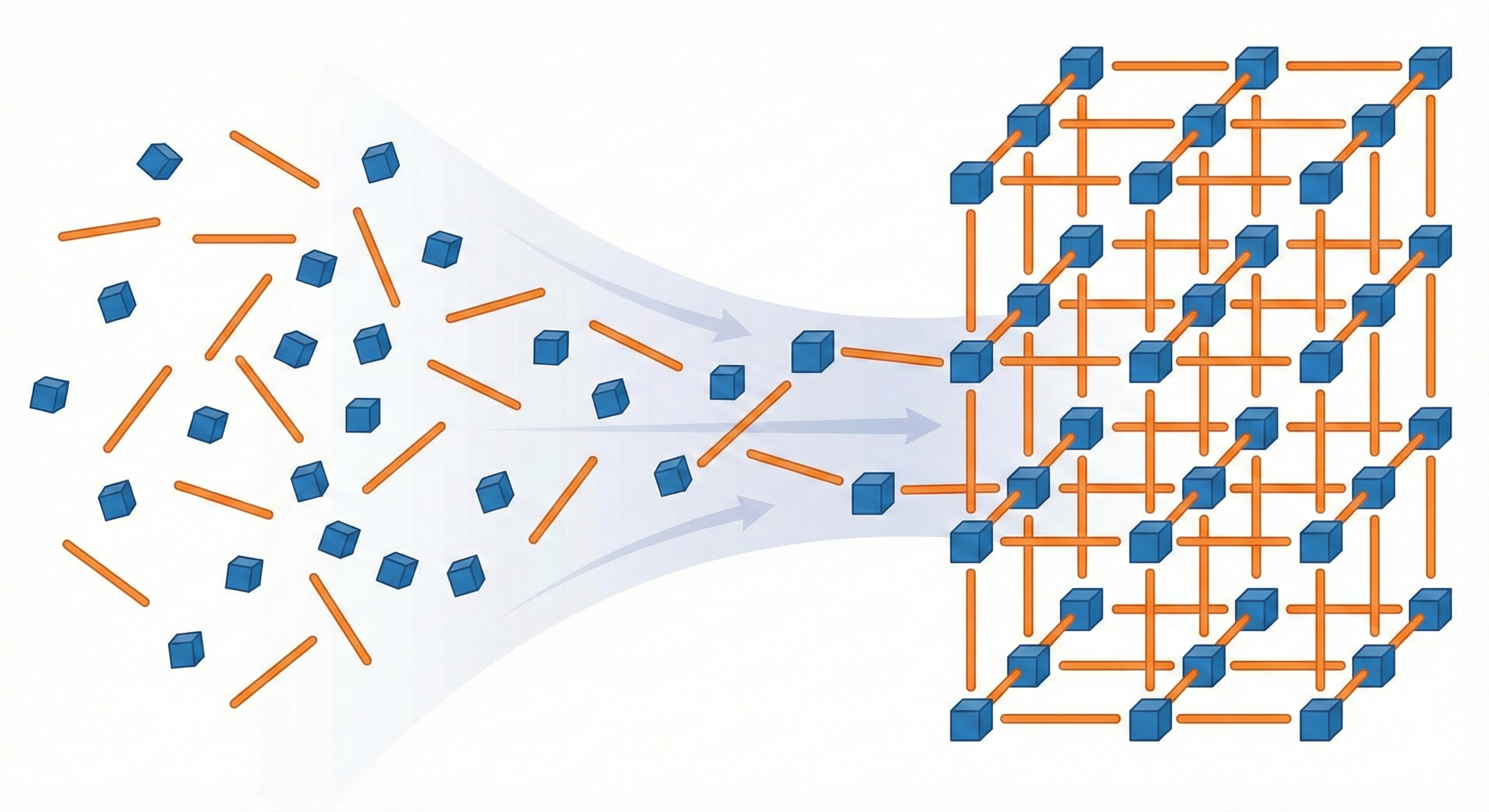
A Riemannian flow matching framework for generating Metal-Organic Framework structures by treating building blocks as …
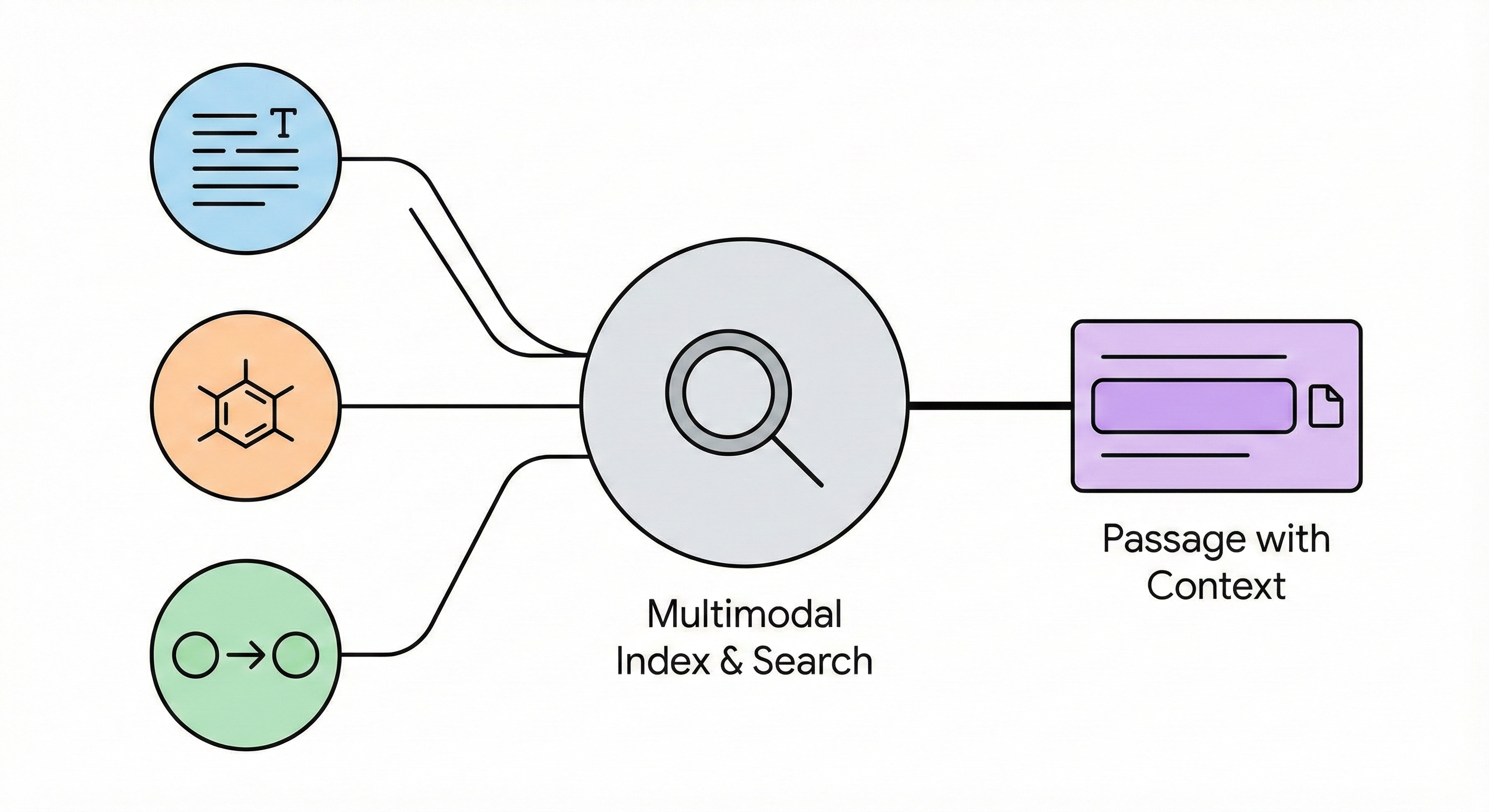
A multimodal search engine that integrates text passages, molecular diagrams, and reaction data to enable passage-level …
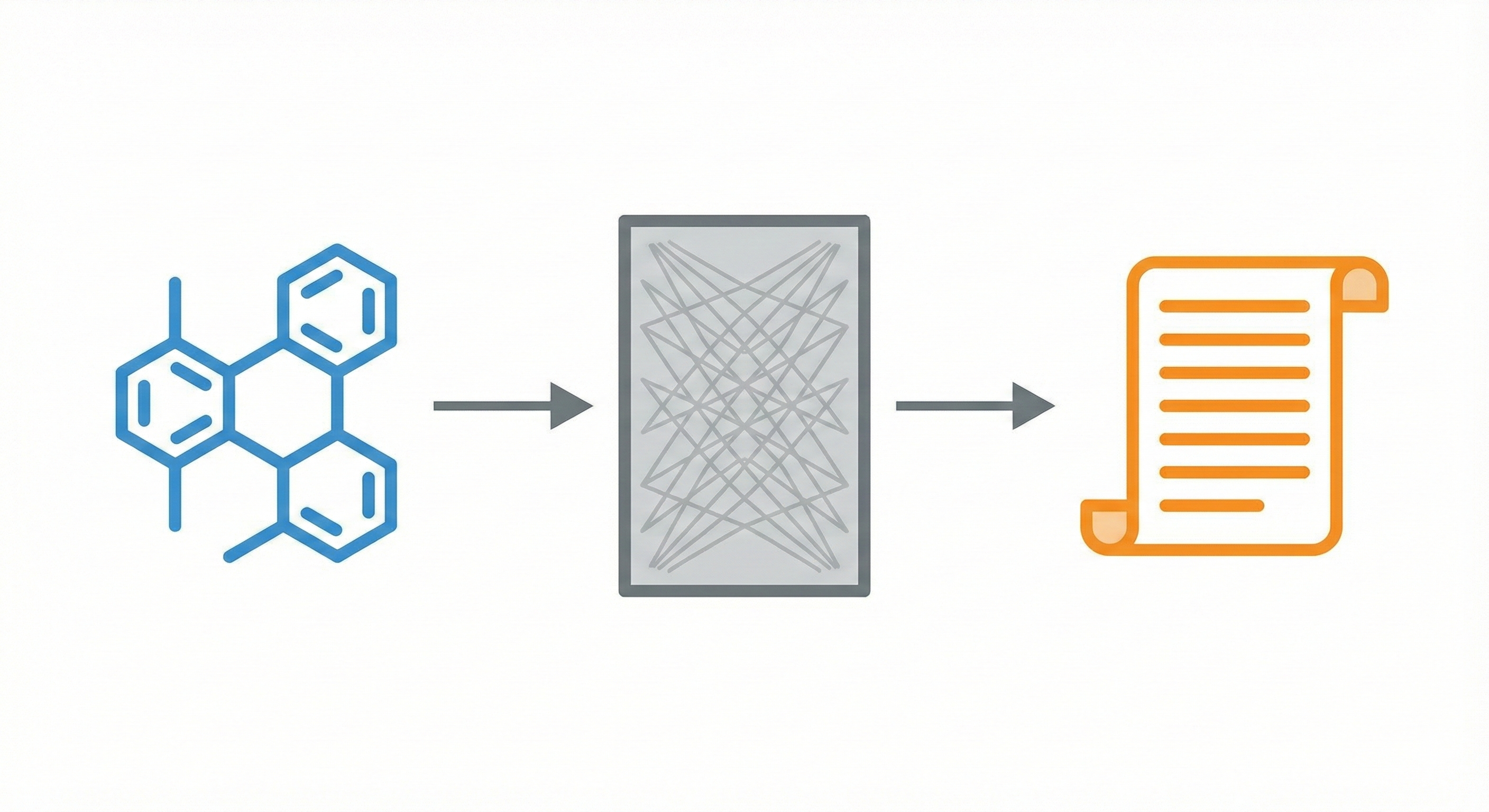
A Transformer-based model for translating SMILES to IUPAC names, trained on ~1 billion molecules, achieving ~0.99 BLEU …

A deep-learning neural machine translation approach to translate between SMILES strings and IUPAC names using the STOUT …
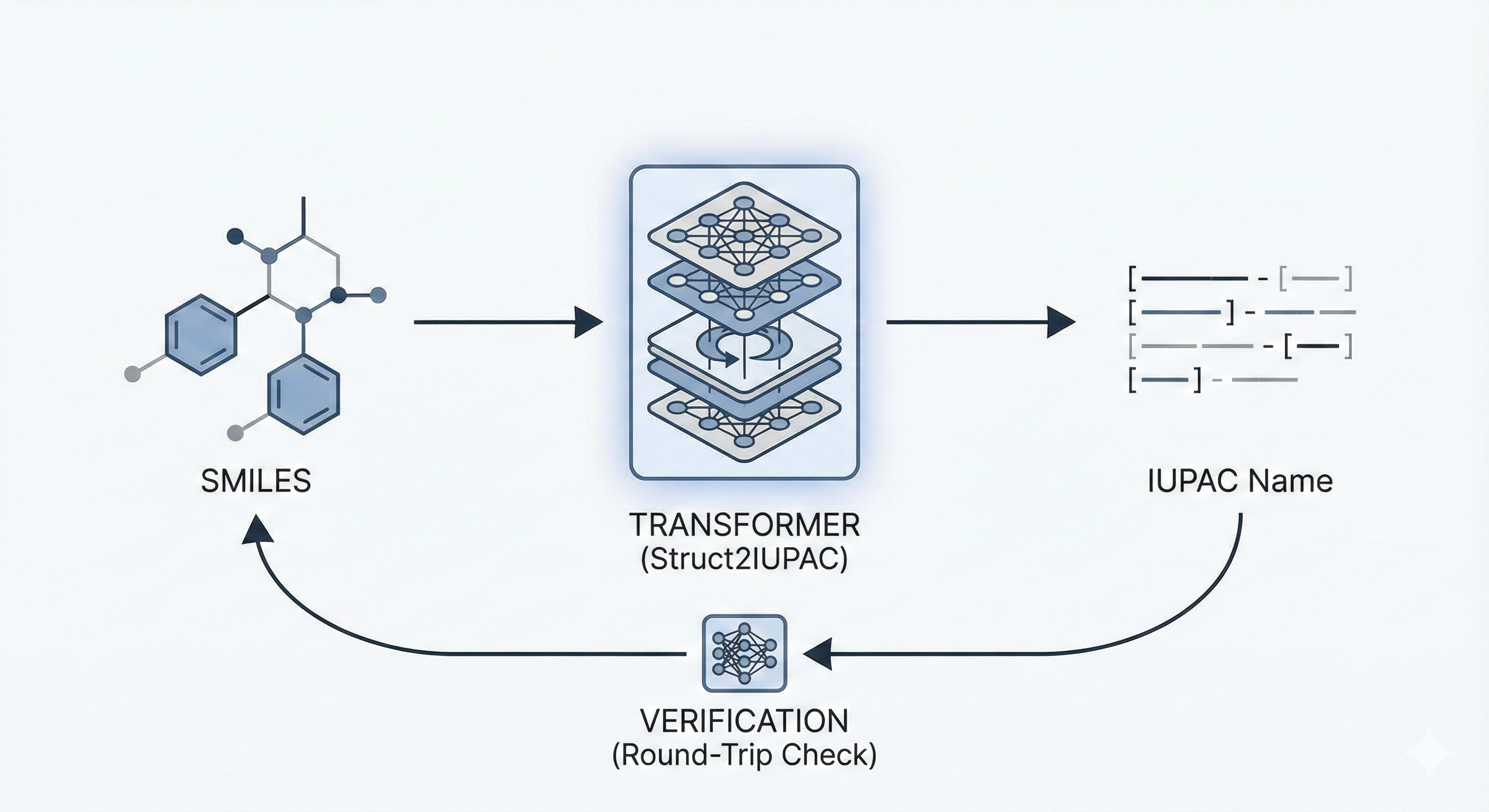
A Transformer-based model for translating between SMILES strings and IUPAC names, trained on 47M PubChem examples, …
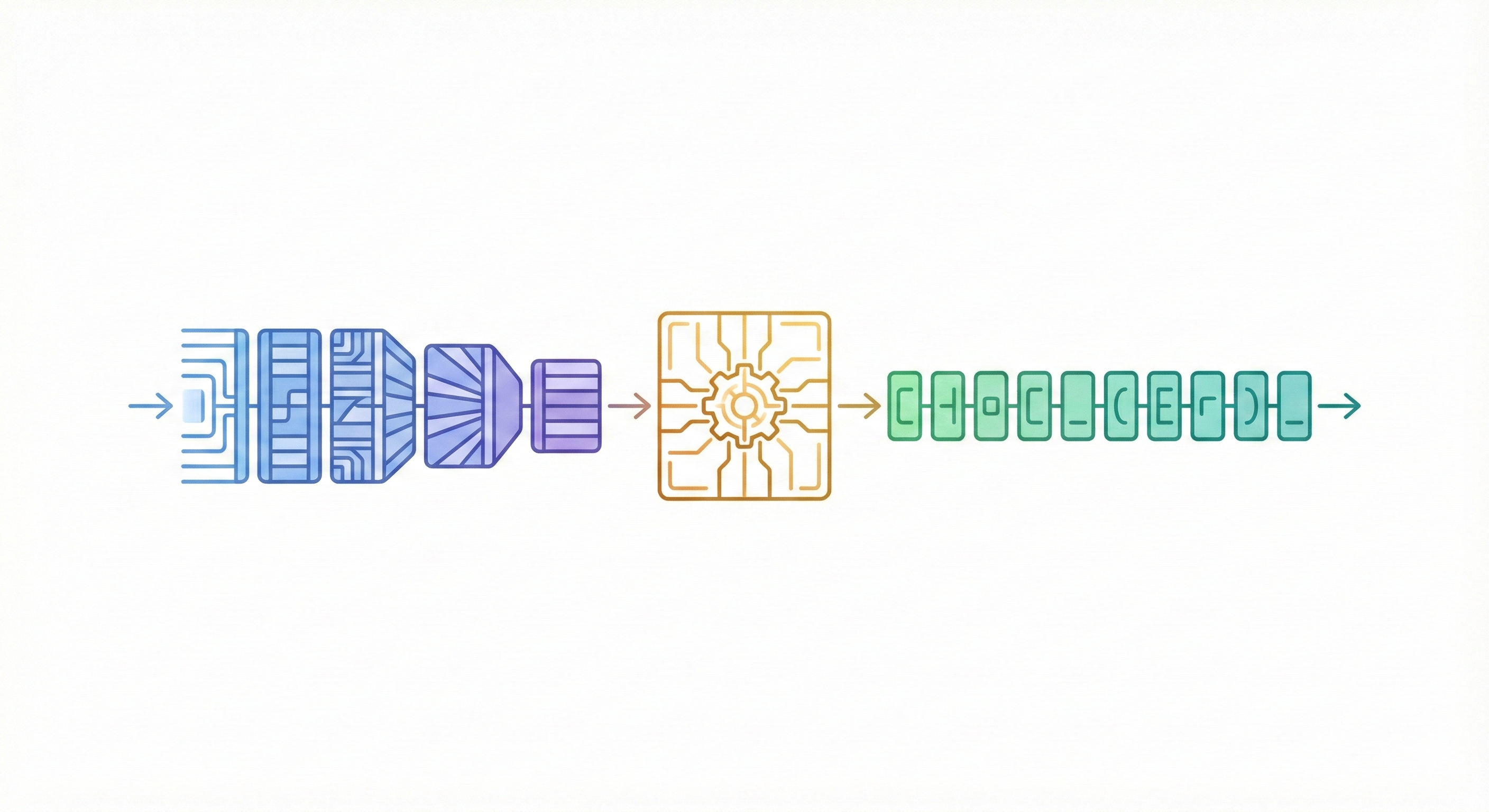
Sequence-to-sequence Transformer translating InChI identifiers to IUPAC names with 91% accuracy on organic compounds.
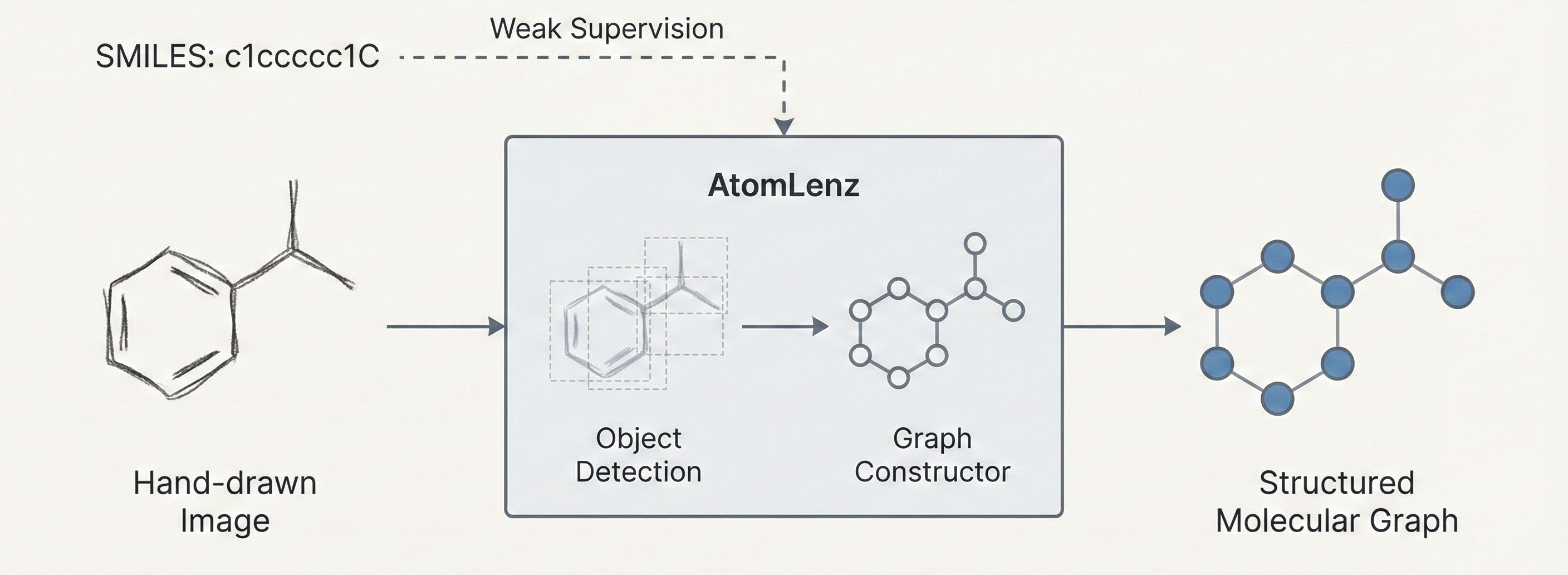
Weakly supervised OCSR framework combining object detection and graph construction to recognize chemical structures from …