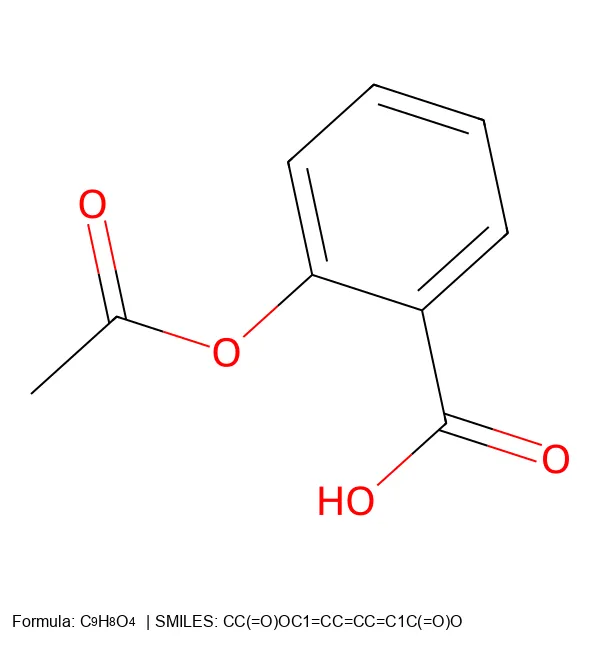
Converting SMILES and SELFIES to 2D Molecular Images
Build a robust Python CLI tool that converts both SMILES and SELFIES notation into publication-quality 2D molecular images, complete with formulas and legends.

Build a robust Python CLI tool that converts both SMILES and SELFIES notation into publication-quality 2D molecular images, complete with formulas and legends.
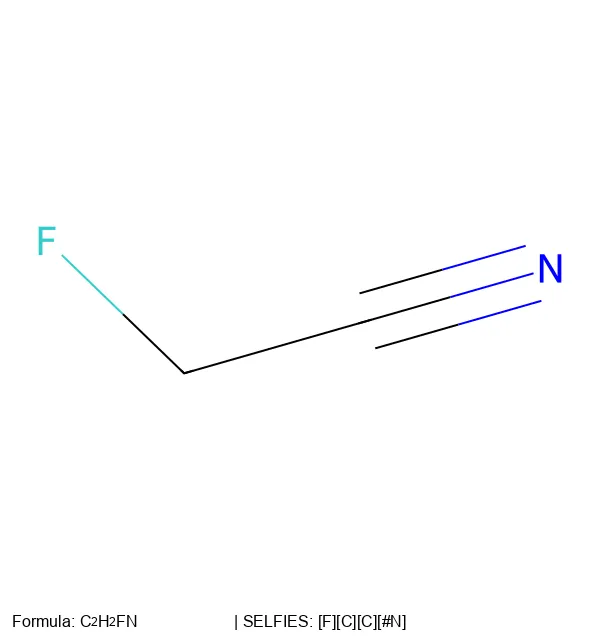
An in-depth overview of SELFIES, the 100% robust molecular string representation designed to overcome SMILES limitations in machine learning, where every possible string (even random ones) decodes to a valid molecule through local operations, customizable valence rules, and graph-based internal representations.
MARCEL provides a comprehensive benchmark for molecular representation learning with 722K+ conformers across four diverse subsets (Drugs-75K, Kraken, EE, BDE), enabling evaluation of conformer ensemble methods for property prediction in drug discovery and catalysis.
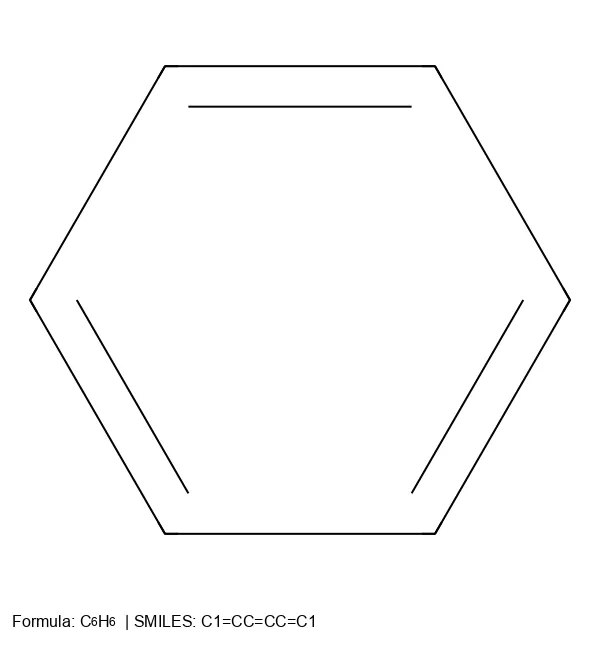
Comprehensive overview of SMILES notation for chemical structures, covering syntax for atoms, bonds, branches, rings, and stereochemistry, plus its key limitations for machine learning.
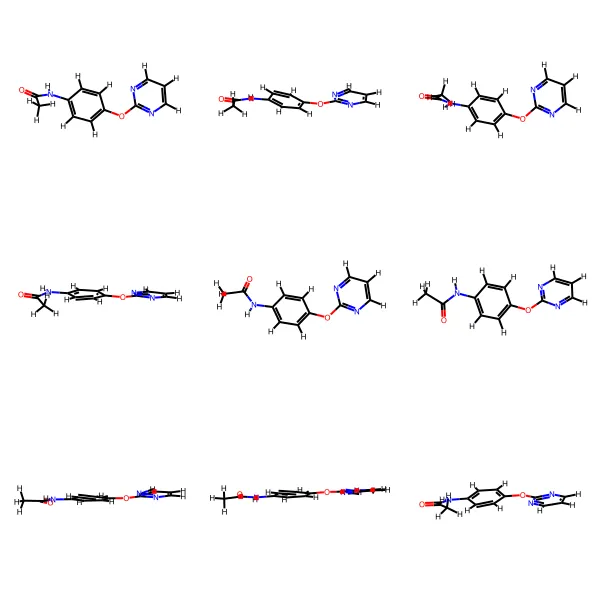
GEOM contains 450k+ molecules with 37M+ conformations, featuring energy annotations from semi-empirical (GFN2-xTB) and DFT methods for property prediction and molecular generation research.
ICLR 2025 paper introducing DenoiseVAE, which learns adaptive, atom-specific noise distributions through a VAE framework to improve denoising-based pre-training for molecular force field prediction, outperforming fixed Gaussian noise approaches on quantum chemistry benchmarks.
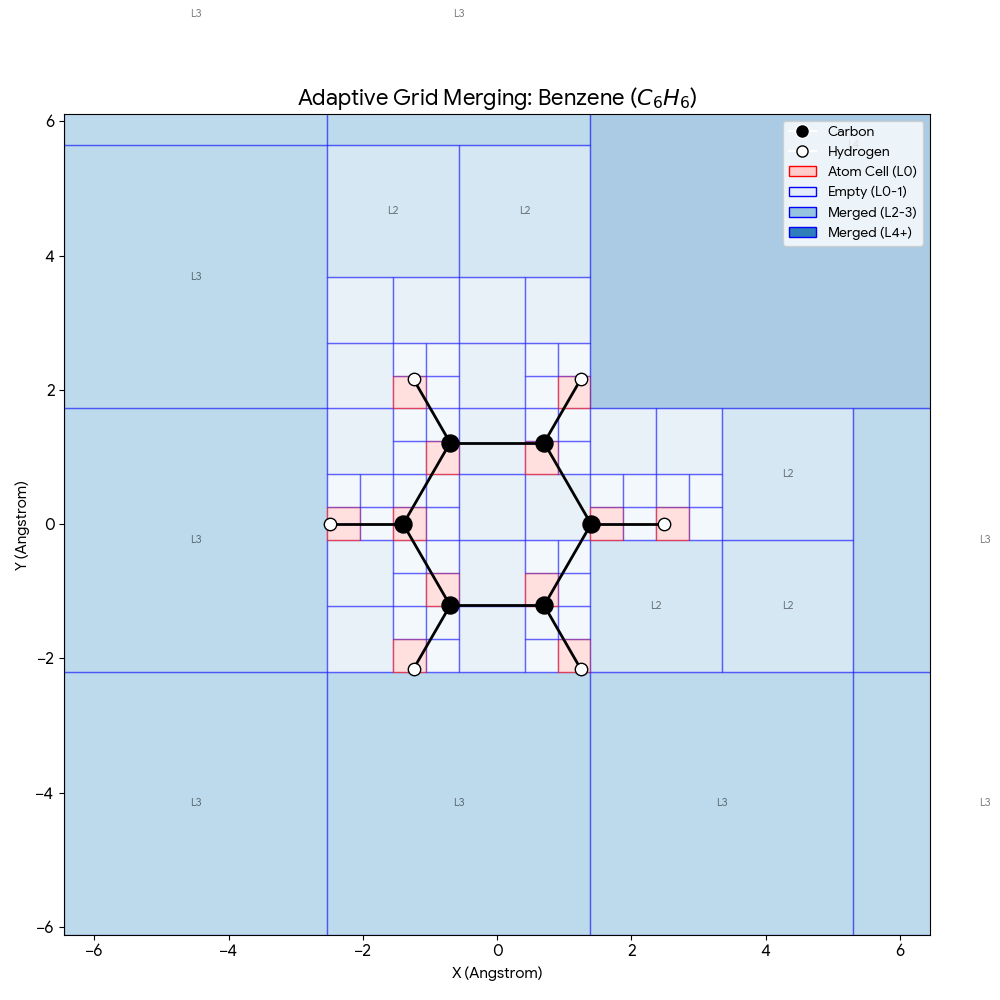
ICML 2025 paper introducing SpaceFormer, a Transformer architecture that challenges the atom-centric paradigm by modeling the continuous 3D space surrounding molecules using adaptive multi-resolution grids, achieving state-of-the-art performance on quantum property prediction benchmarks.
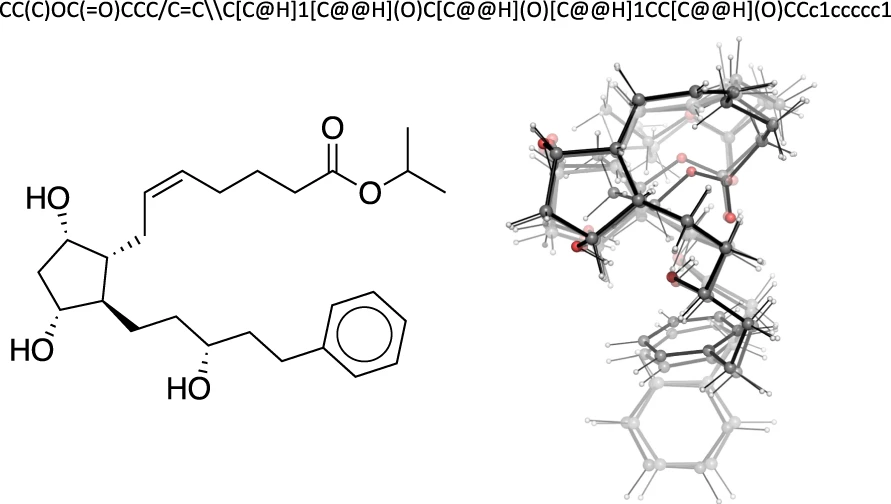
Get a practical overview of the GEOM dataset and learn how it’s advancing 3D molecular machine learning by bridging static graphs and dynamic reality.
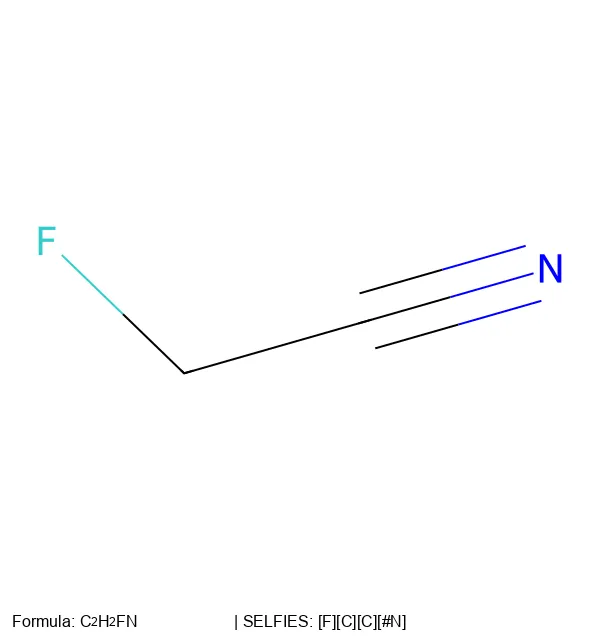
A provocative 2024 Nature Machine Intelligence paper challenging the assumption that invalid SMILES are failures, showing empirically that the ability to generate invalid outputs actually improves chemical language model performance by enabling quality filtering and providing richer training signals.
An end-to-end data factory for molecular machine learning that transforms raw chemical formulas (e.g., C6H14) into labeled 3D conformer datasets, using MAYGEN for structural isomer enumeration, RDKit for 3D embedding, and physics-based featurization to address data scarcity in computational drug discovery.

Can mathematical signatures capture molecular shape? We test whether Coulomb matrix eigenvalues can distinguish alkane constitutional isomers, from unsupervised clustering failures to supervised learning successes.
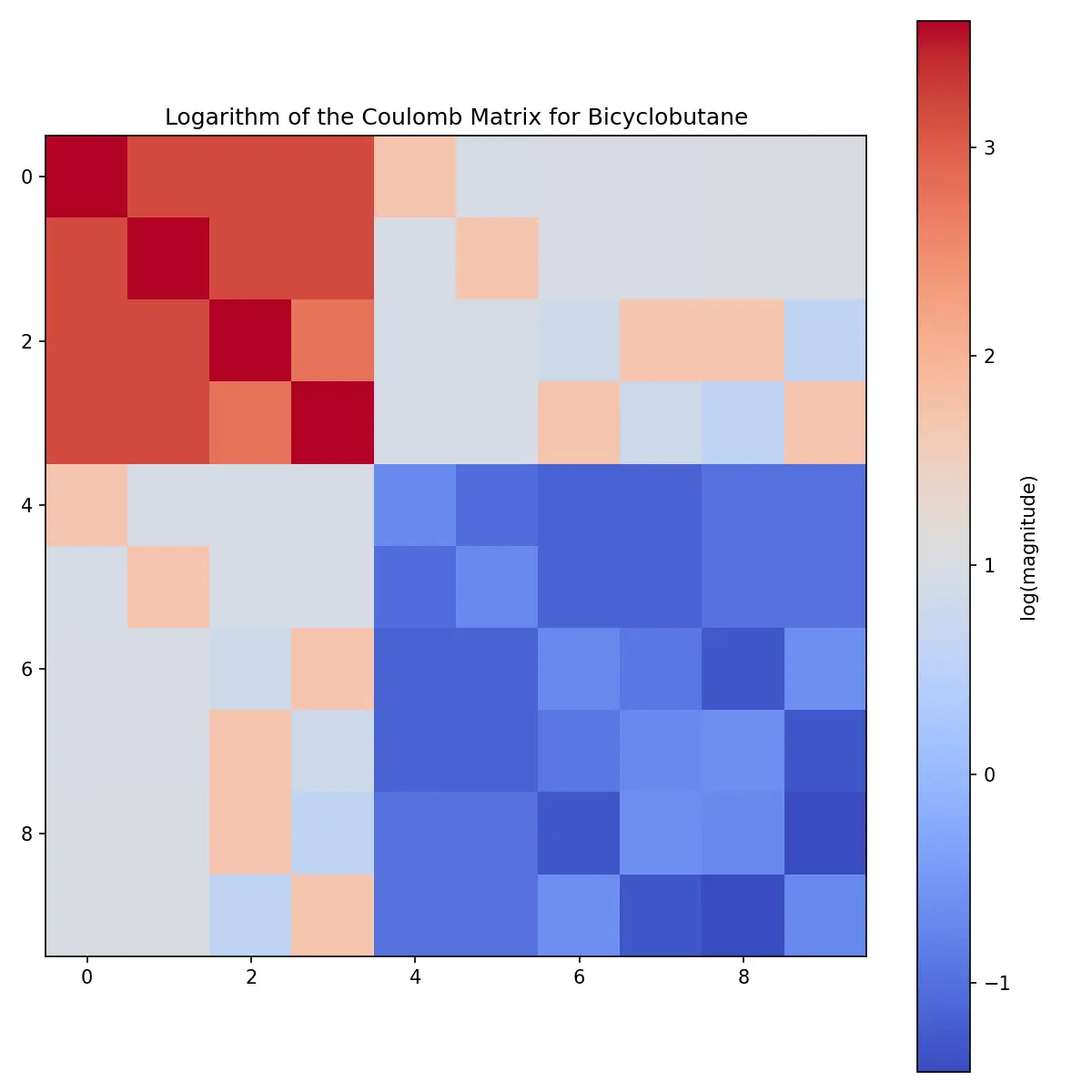
A practical introduction to Coulomb matrices: how they transform molecular 3D structures into ML features, complete with Python examples and honest assessment of their limitations.