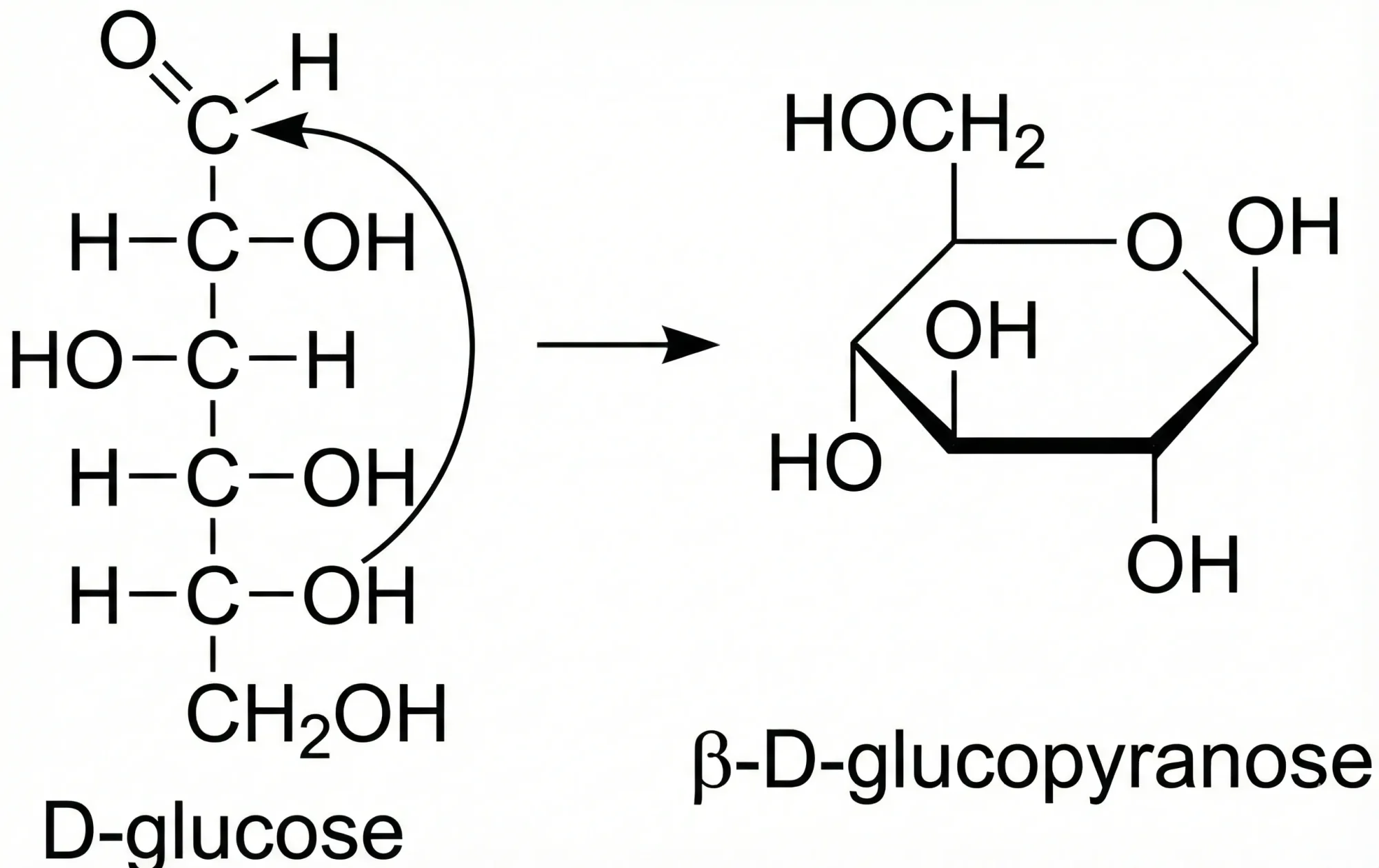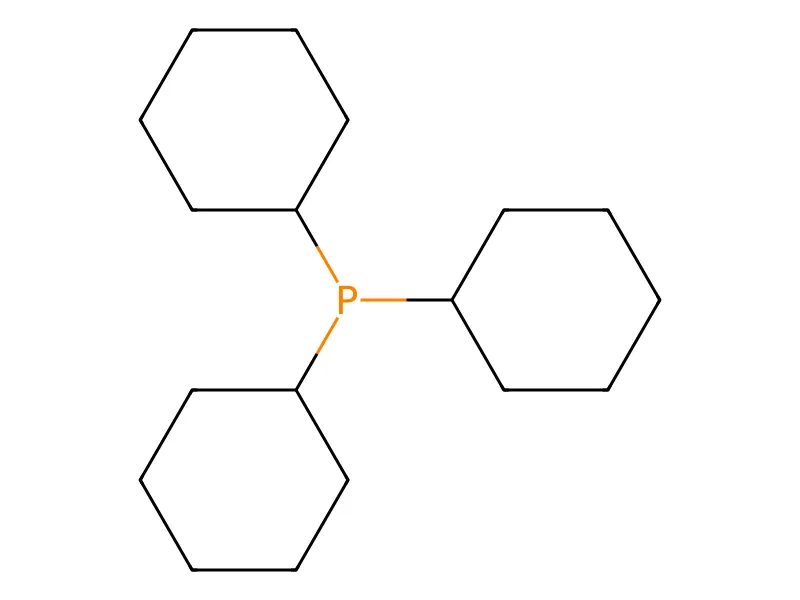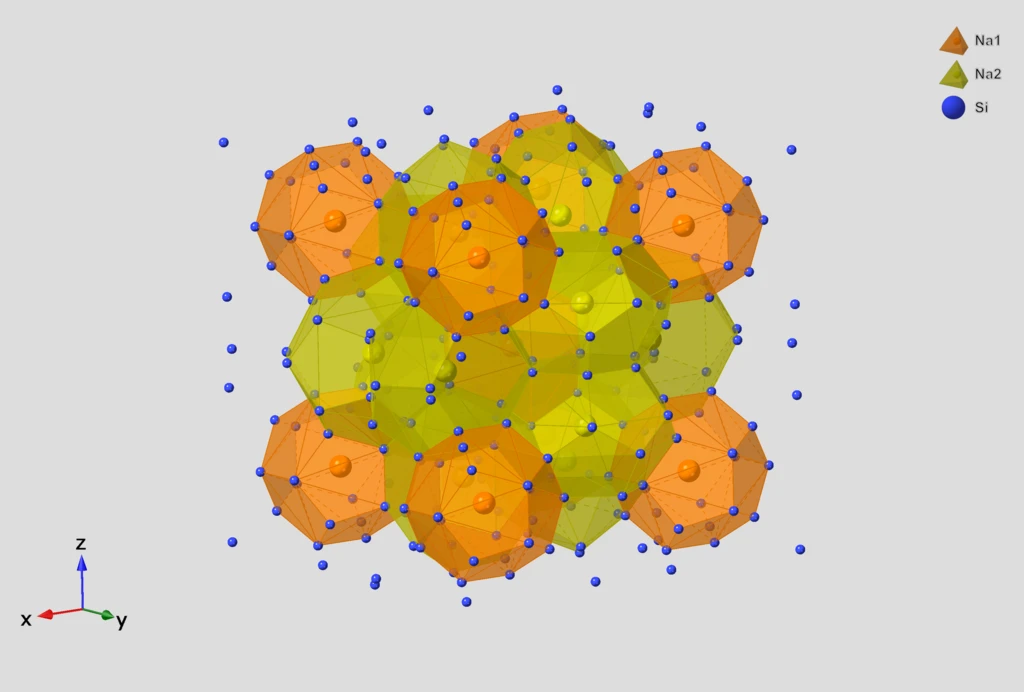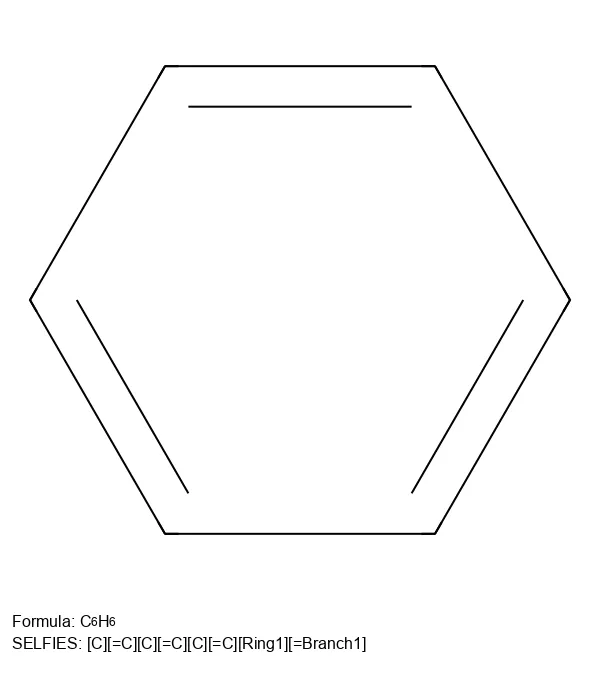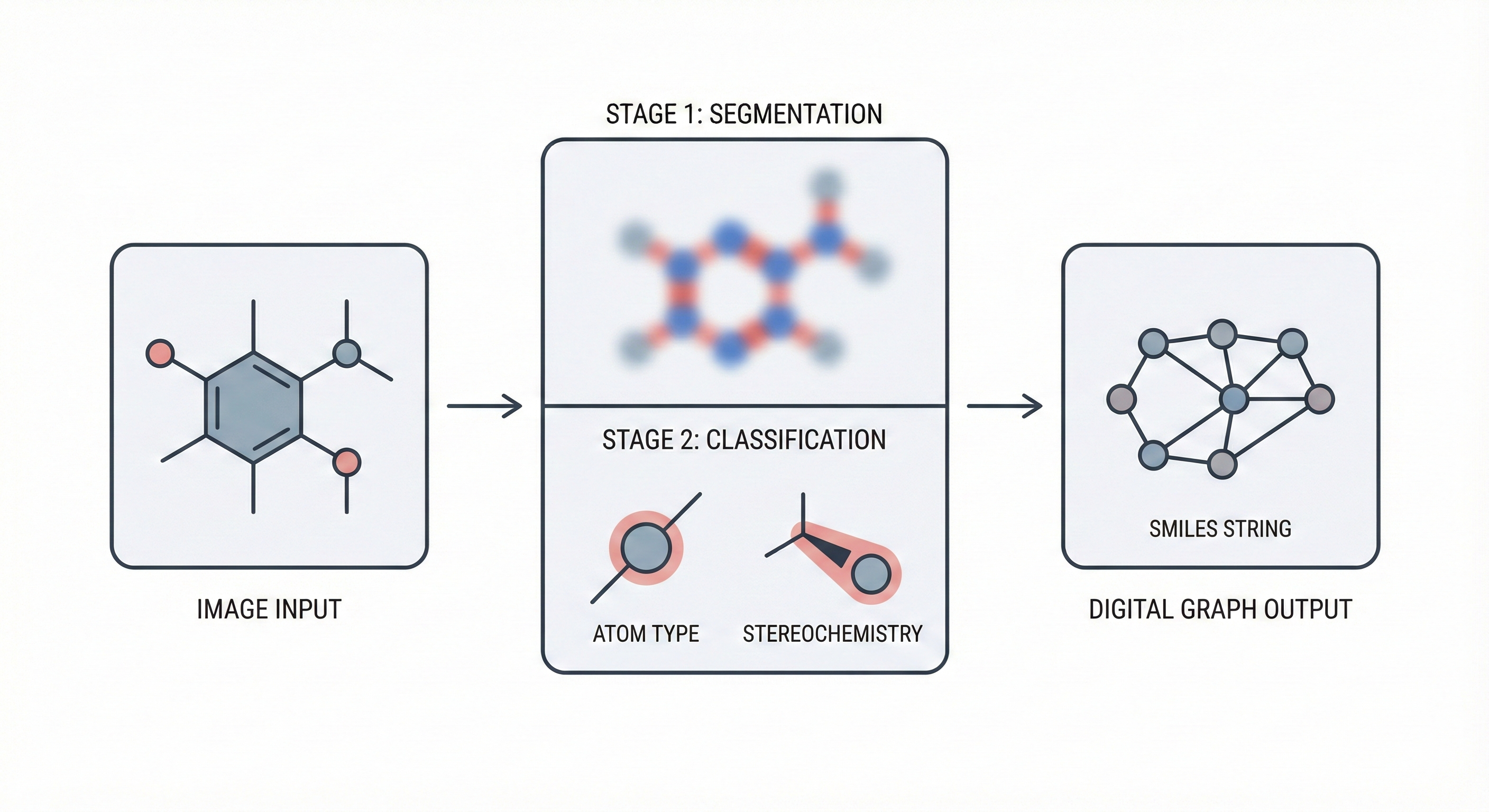
ChemGrapher: Deep Learning for Chemical OCR
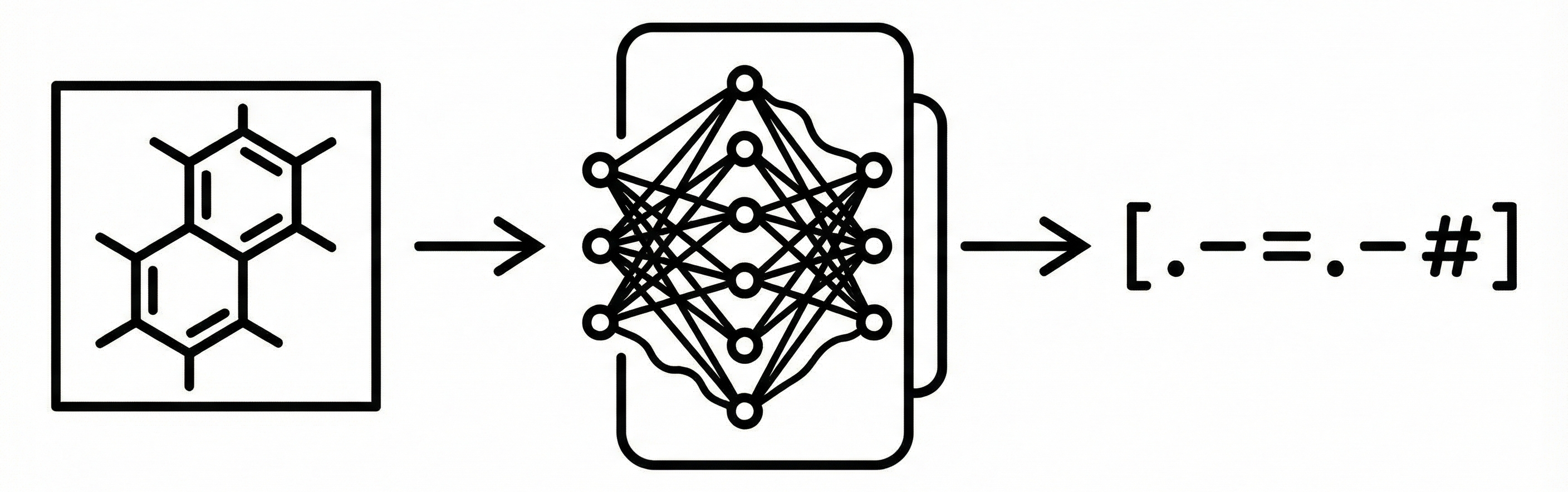
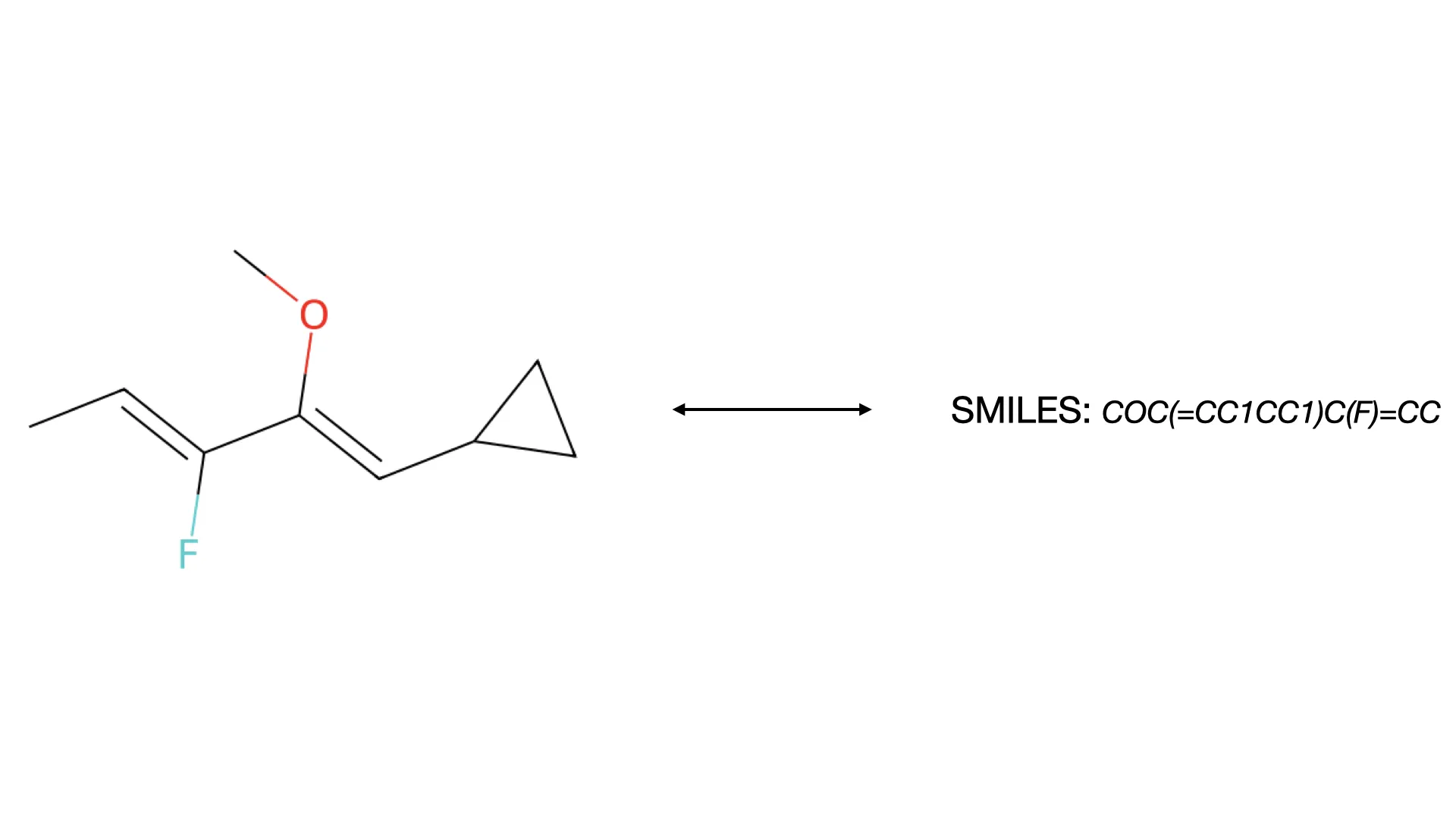
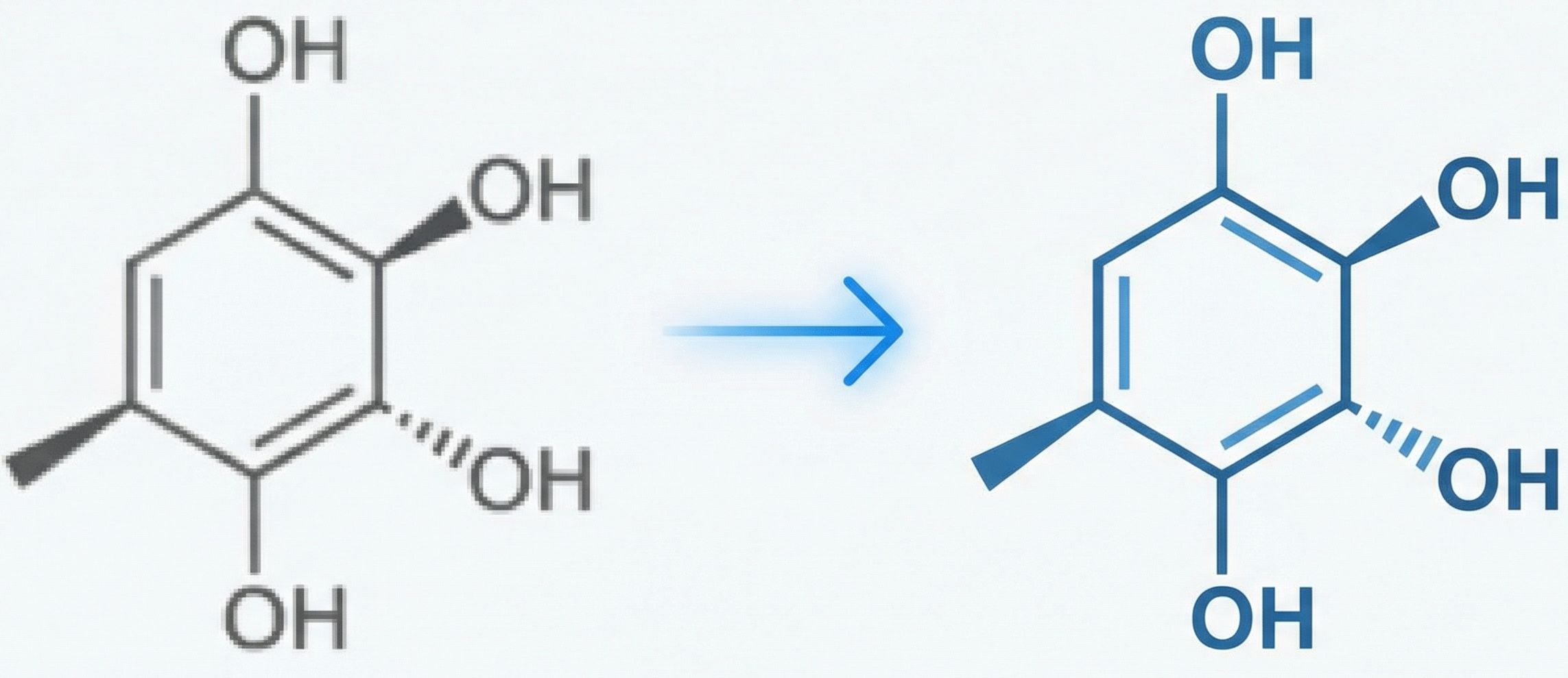
ChemGrapher replaces rule-based chemical OCR with a deep learning pipeline using semantic segmentation to identify atom and bond candidates, followed by specialized classification networks to resolve stereochemistry and bond multiplicity, significantly outperforming OSRA.