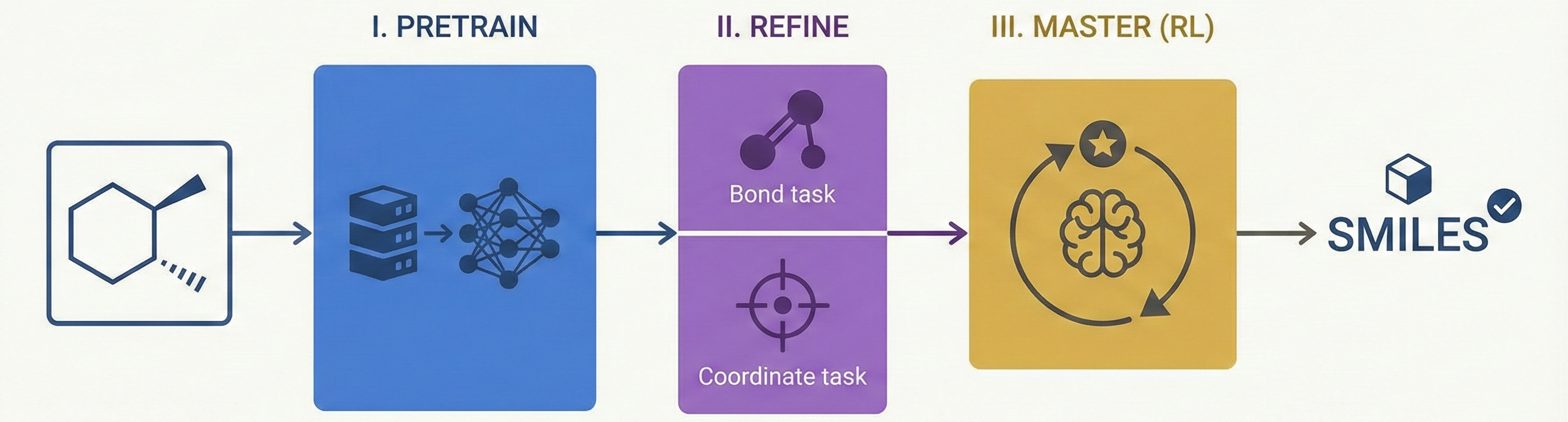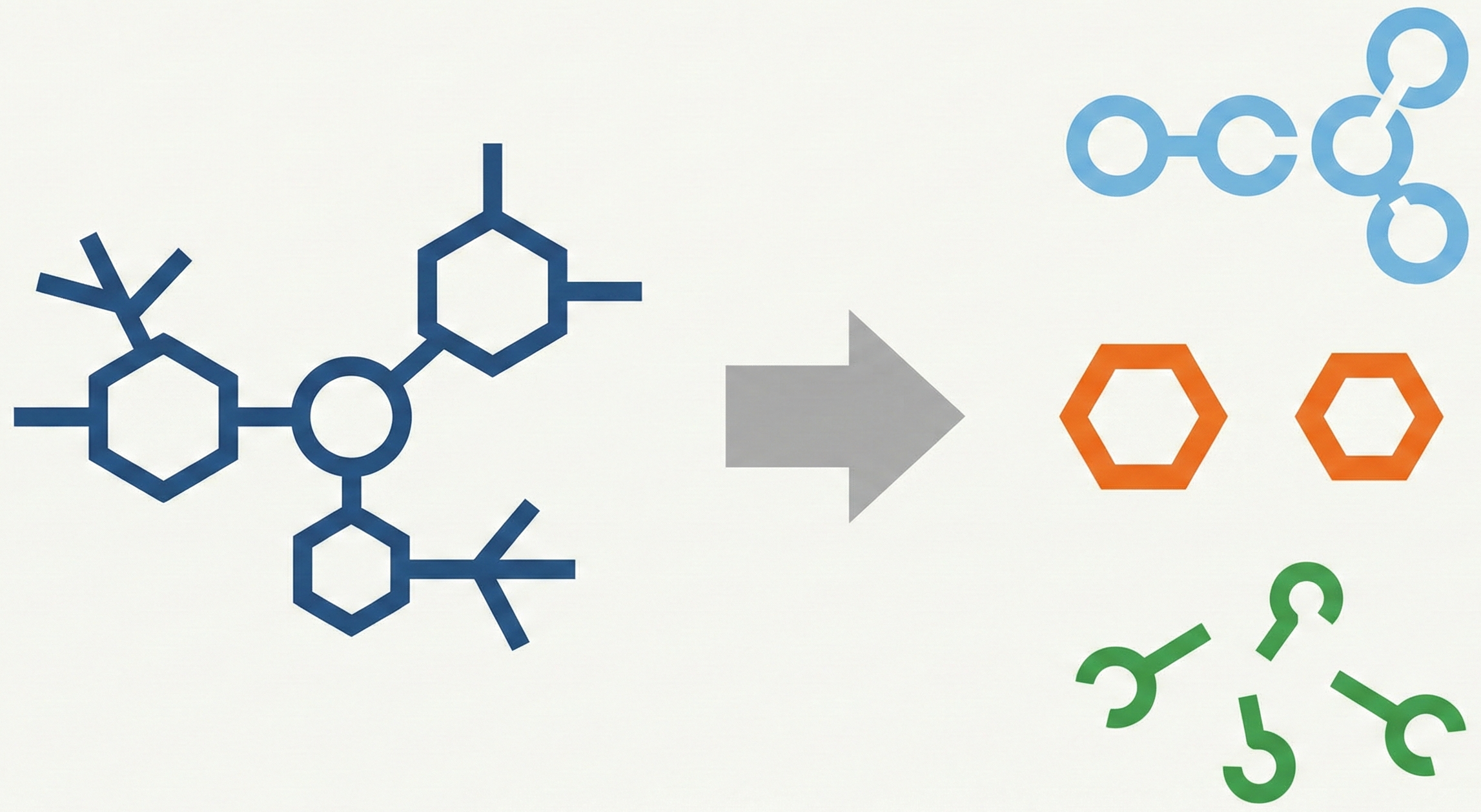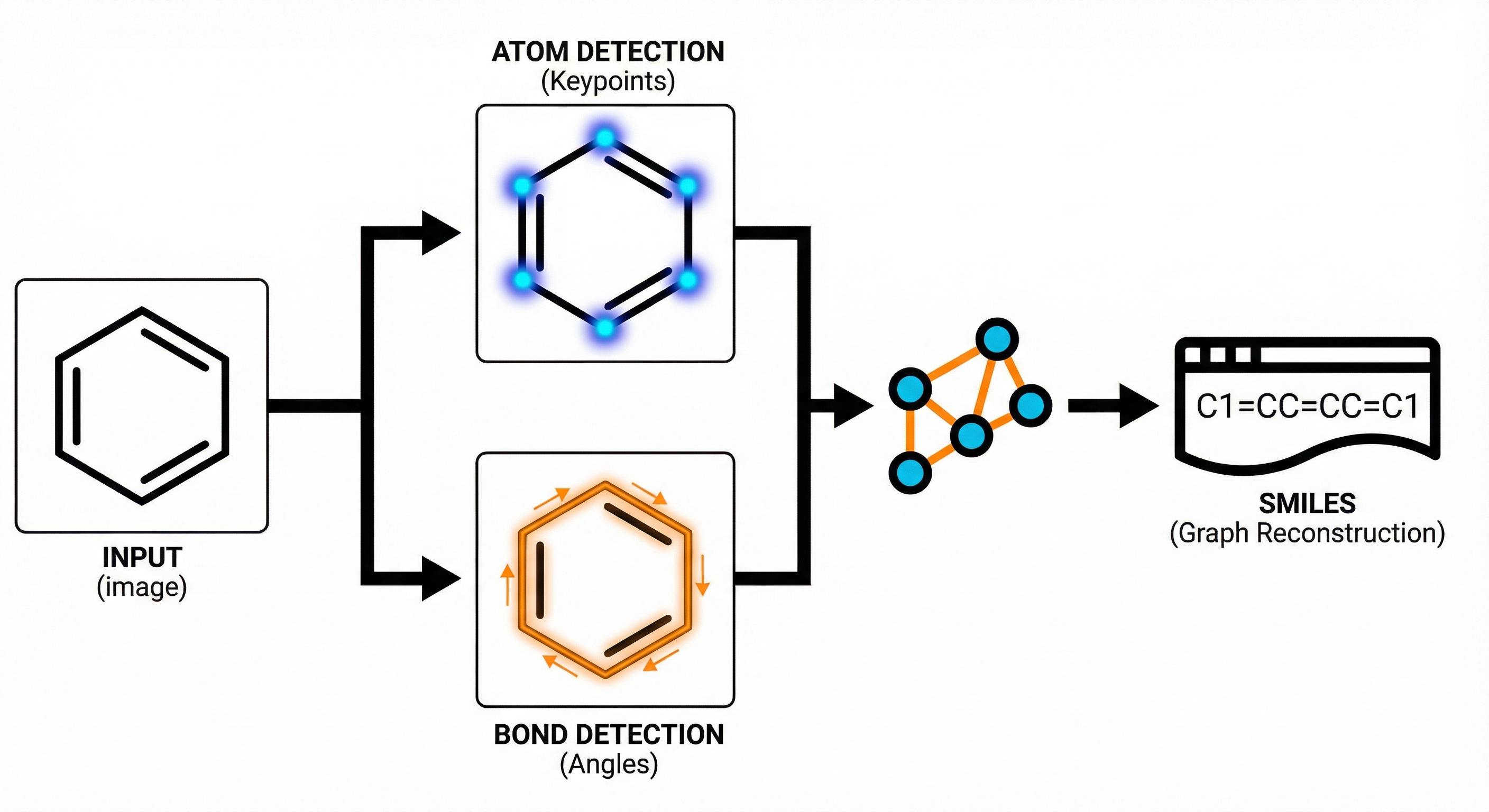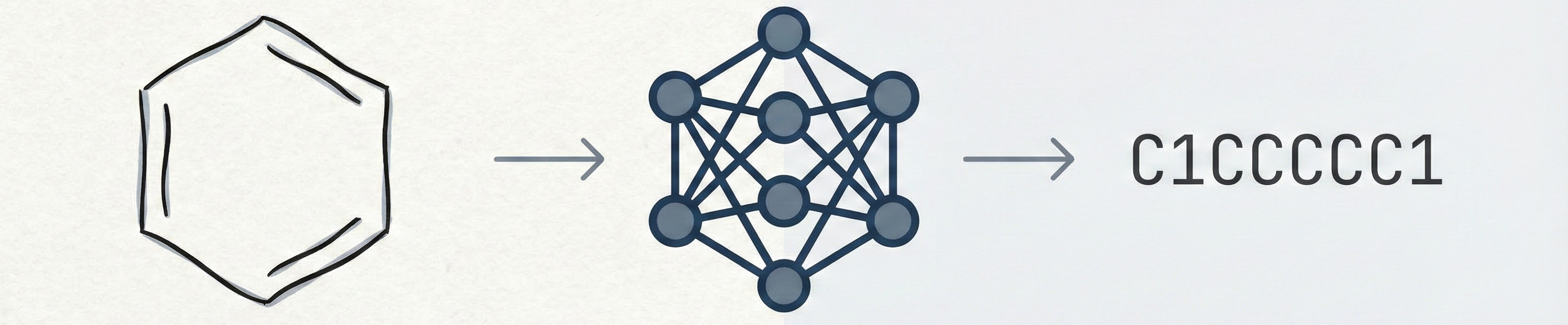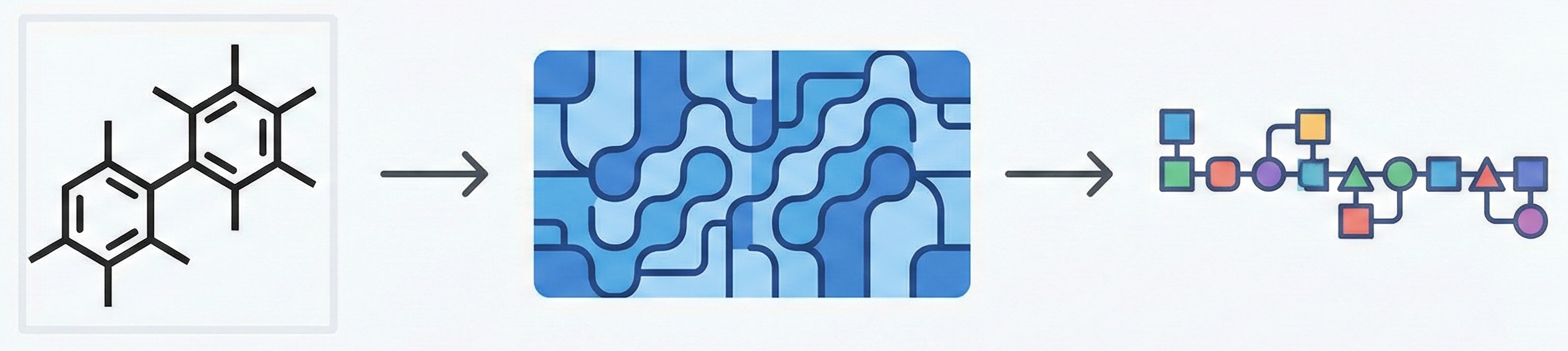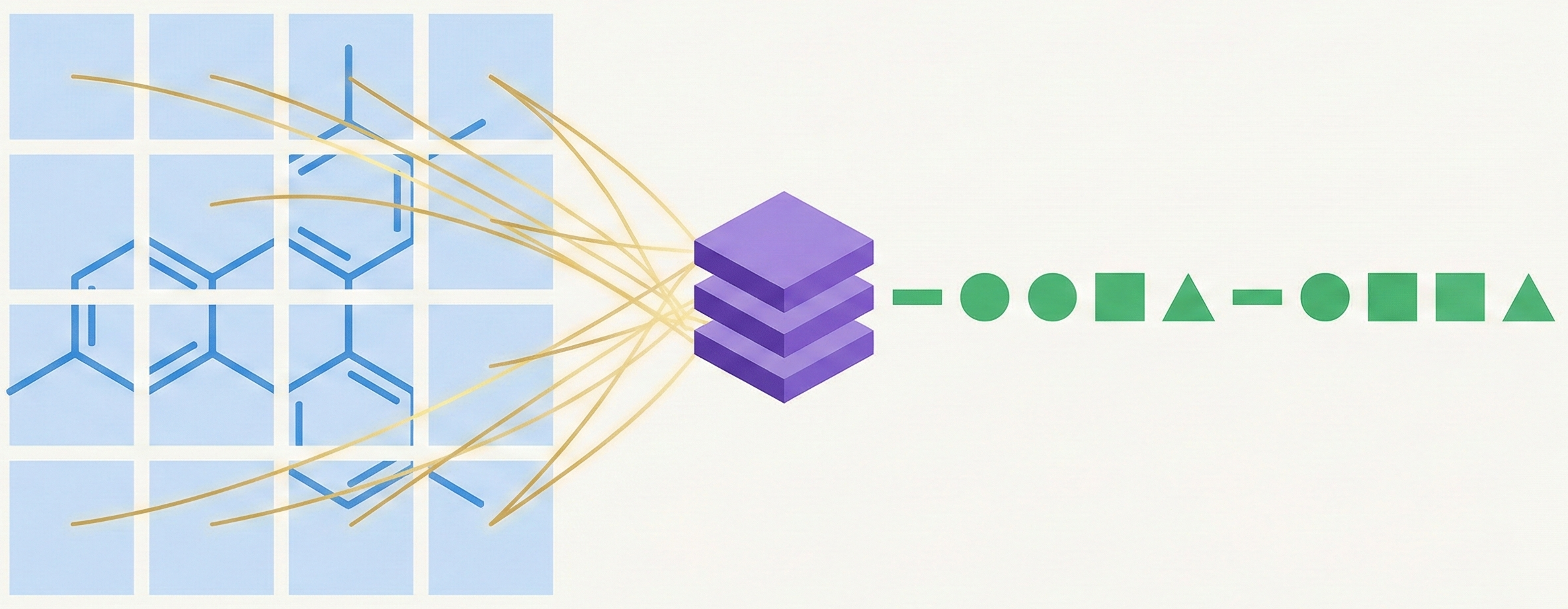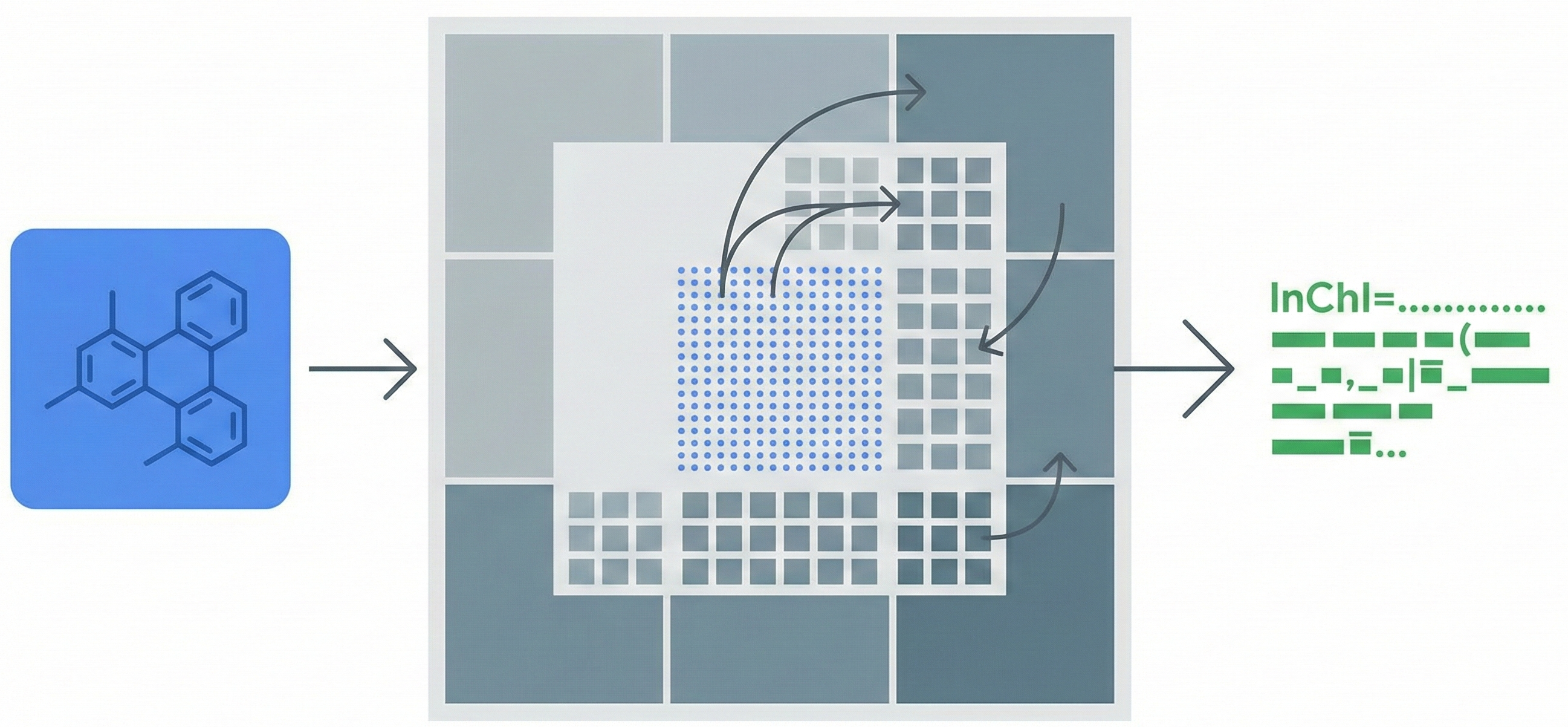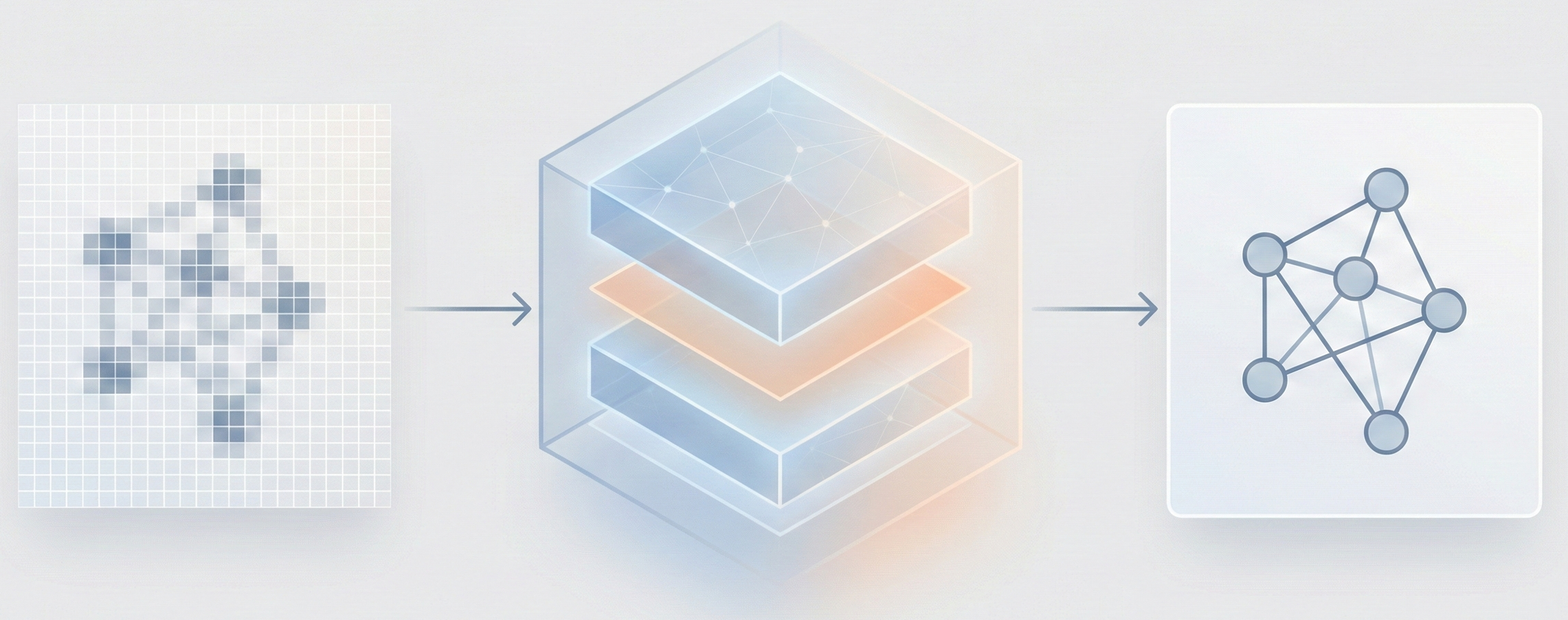
MarkushGrapher: Multi-modal Markush Structure Recognition
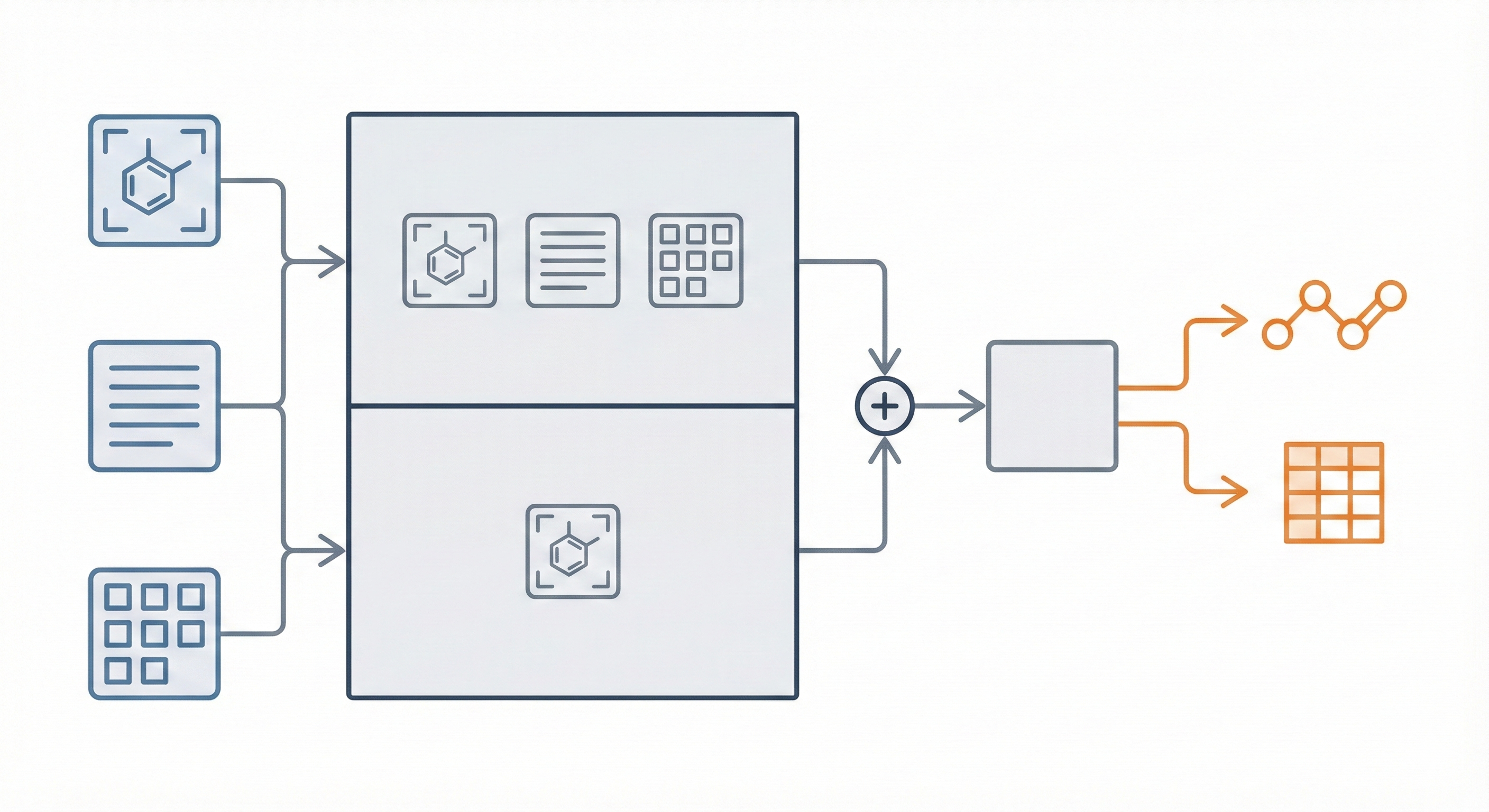
This paper introduces a novel multi-modal approach for extracting chemical Markush structures from patents, combining a Vision-Text-Layout encoder with a specialized chemical vision encoder. It addresses the lack of training data with a robust synthetic generation pipeline and introduces M2S, a new real-world benchmark.