
Implementing the Müller-Brown Potential in PyTorch
Guide to implementing the Müller-Brown potential in PyTorch, comparing analytical vs automatic differentiation with …

Guide to implementing the Müller-Brown potential in PyTorch, comparing analytical vs automatic differentiation with …

Langevin dynamics simulation showing particle motion in the deep reactant minimum (Basin MA) of the Müller-Brown …

Langevin dynamics simulation showing particle motion in the product minimum (Basin MB) of the Müller-Brown potential …

GPU-accelerated PyTorch framework for the Müller-Brown potential with JIT compilation, Langevin dynamics, and …

Extended Langevin dynamics simulation showing particle transitions between different basins of the Müller-Brown …
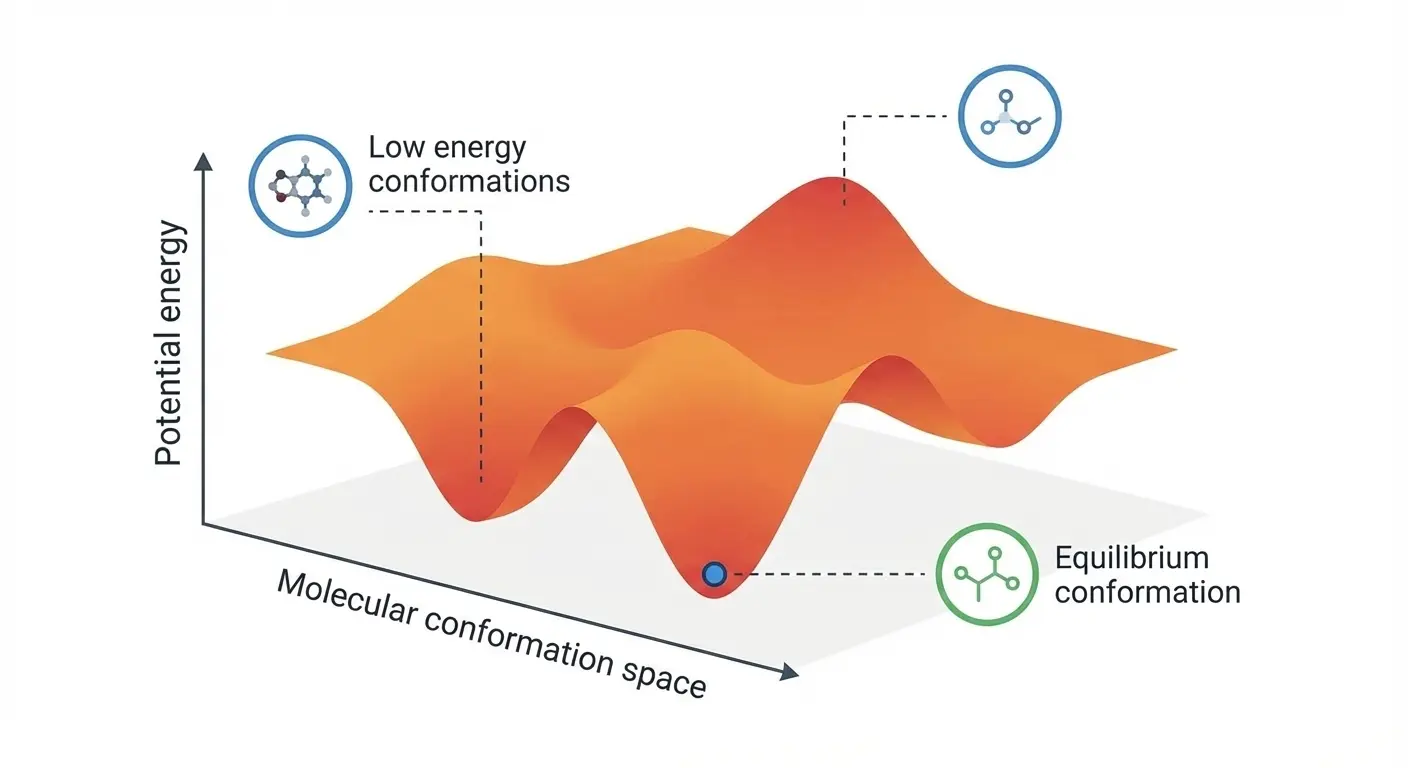
Liu et al.'s ICLR 2025 paper introducing DenoiseVAE, which learns adaptive, atom-specific noise for better molecular …
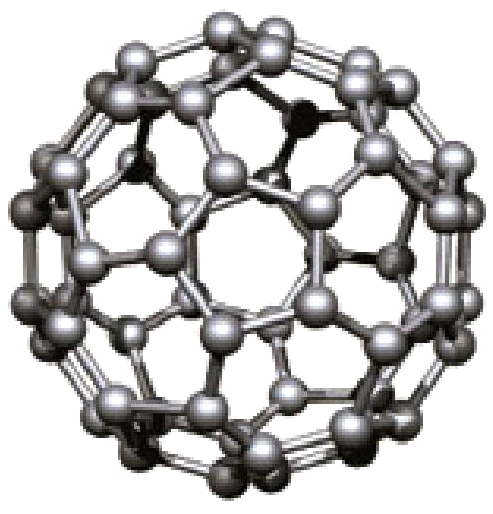
Bigi et al. critique non-conservative force models in ML potentials, showing their simulation failures and proposing …

Fu et al. propose energy conservation as a key MLIP diagnostic and introduce eSEN, bridging test accuracy and real …
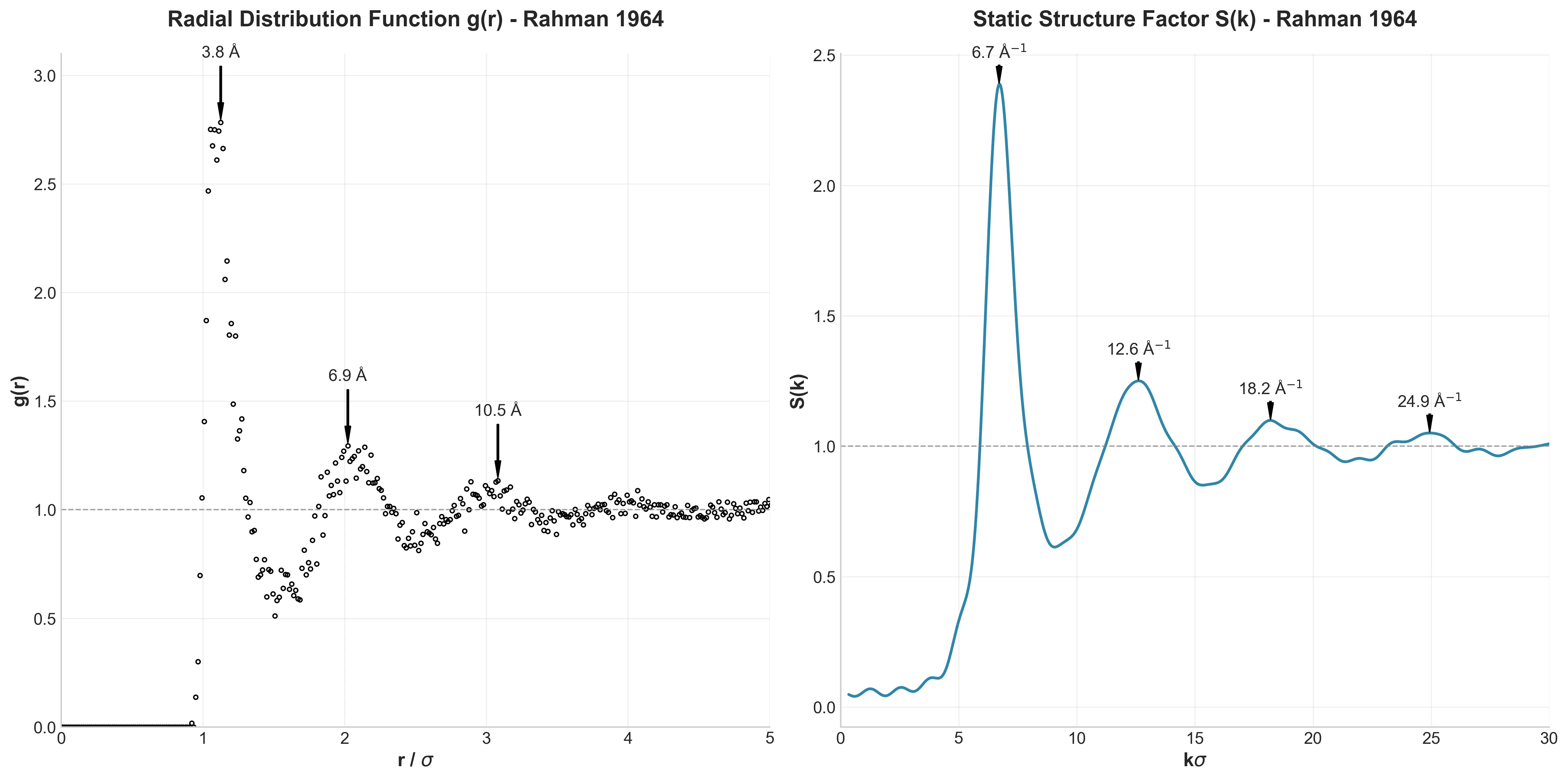
LAMMPS molecular dynamics simulation of liquid argon demonstrating fundamental liquid-state behavior and molecular …
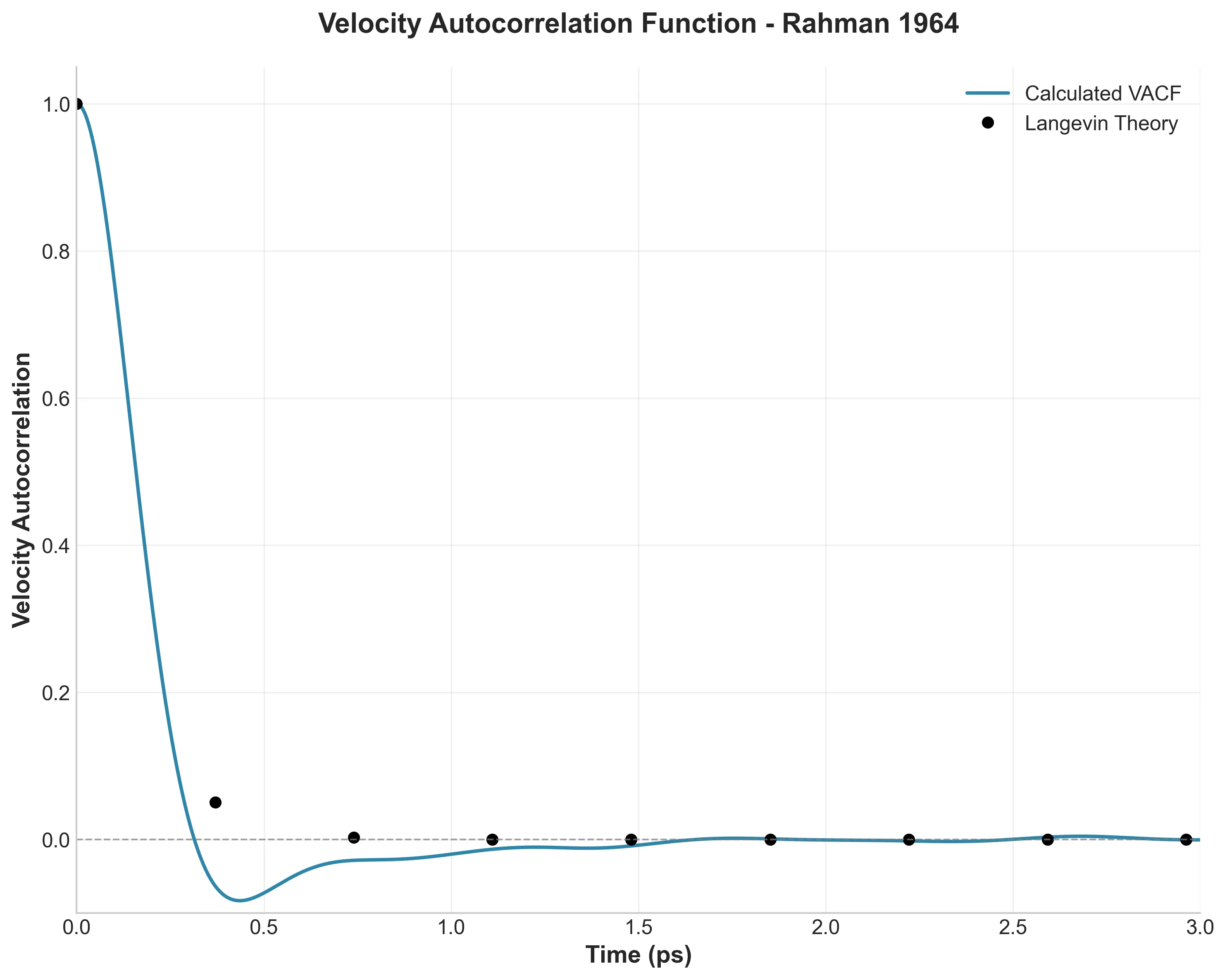
A high-fidelity replication of foundational molecular dynamics using modern software engineering practices: caching, …

How I used modern software engineering (caching, vectorization, and dependency locking) to reproduce a 60-year-old …
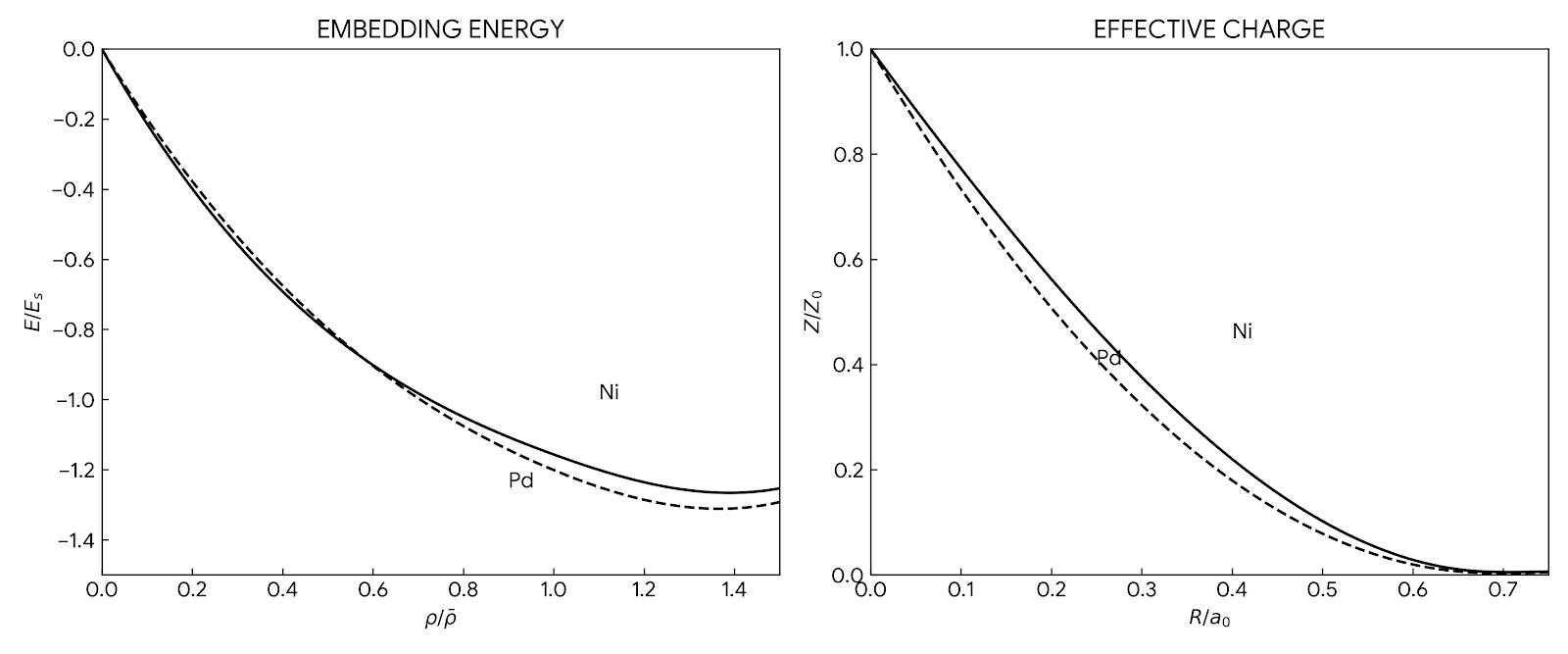
Daw and Baskes's foundational 1984 paper introducing the Embedded-Atom Method (EAM), a many-body potential for metal …