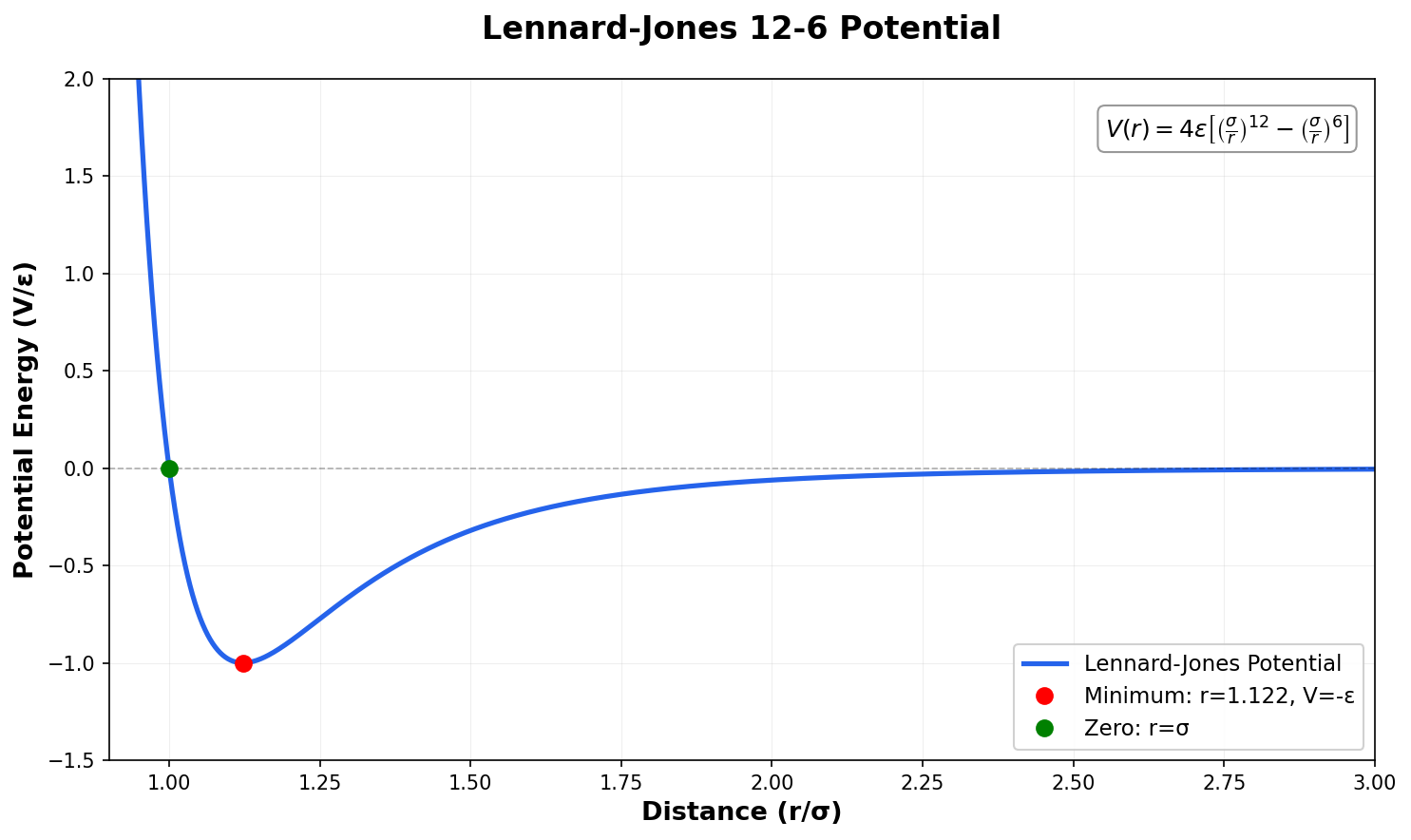
Dynamical Corrections to TST for Surface Diffusion
Application of dynamical corrections formalism to TST for LJ surface diffusion, revealing bounce-back recrossings at low …

Application of dynamical corrections formalism to TST for LJ surface diffusion, revealing bounce-back recrossings at low …
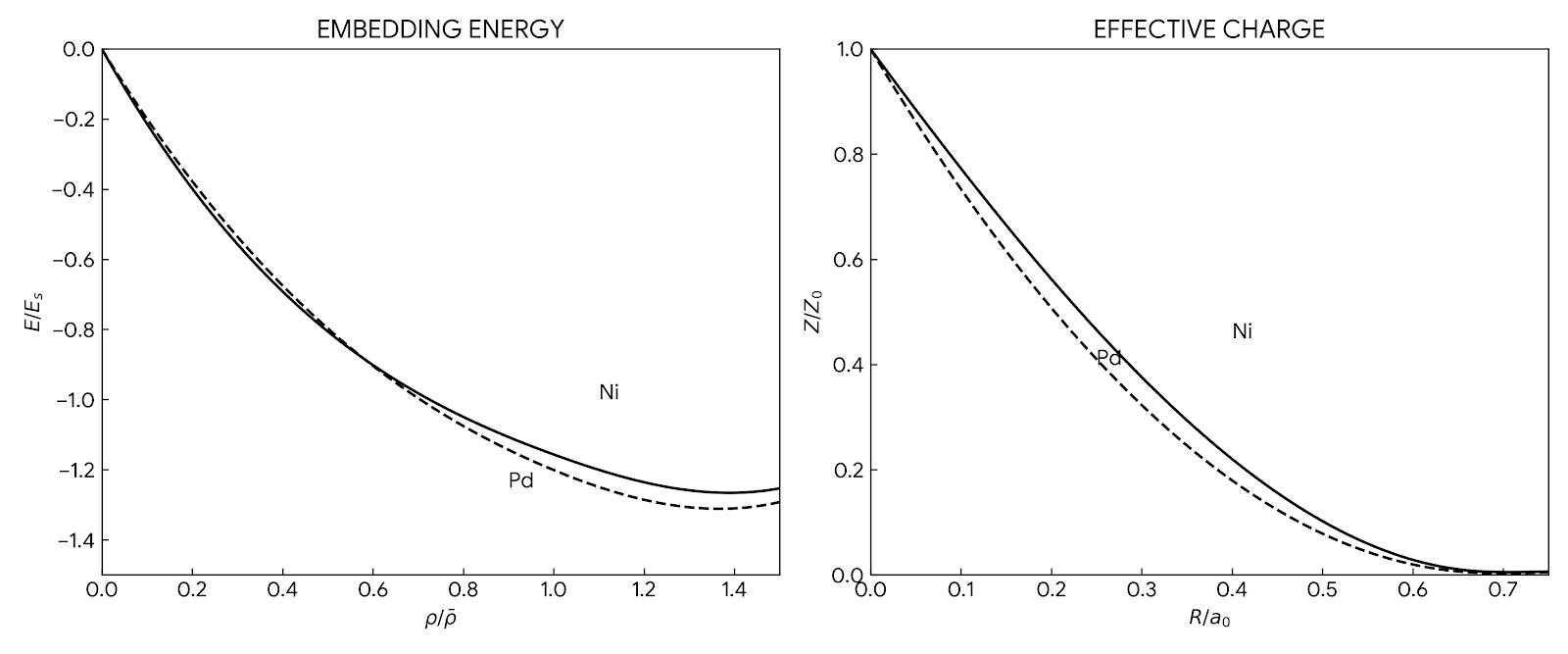
Comprehensive user guide for the Embedded-Atom Method (EAM), covering theory, potential fitting, and applications to …

Comprehensive 1993 review of the Embedded-Atom Method (EAM), covering theory, parameterization, and applications to …
Theoretical model using coupled differential equations to explain CO oxidation oscillations via surface phase …
Molecular dynamics simulation of Iridium surface diffusion confirming atomic exchange mechanisms using EAM and many-body …
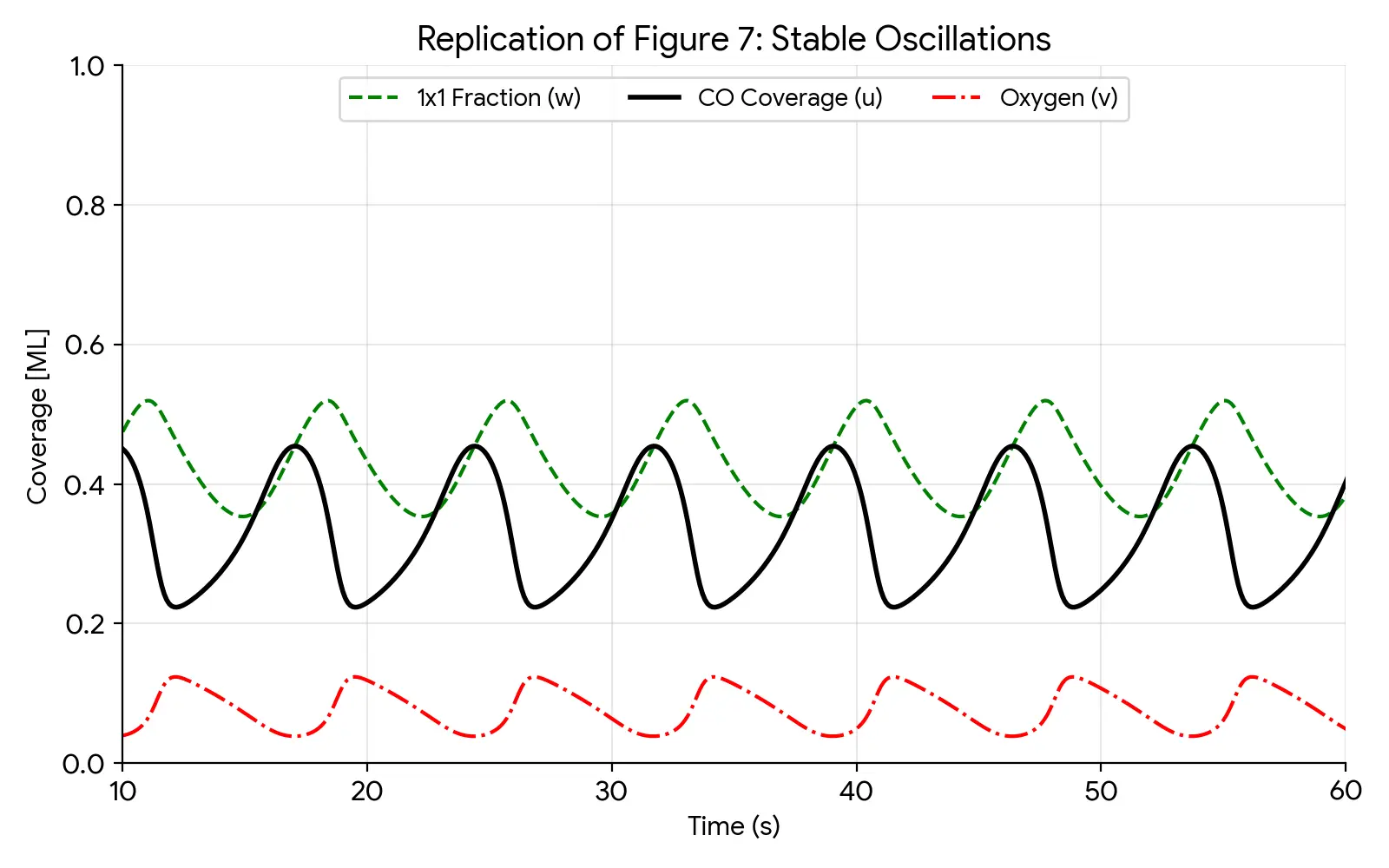
A seminal kinetic model using coupled ODEs to explain temporal self-organization and mixed-mode oscillations on platinum …

In situ XRD validation of the oxide model driving kinetic rate oscillations in high-pressure CO oxidation on supported …
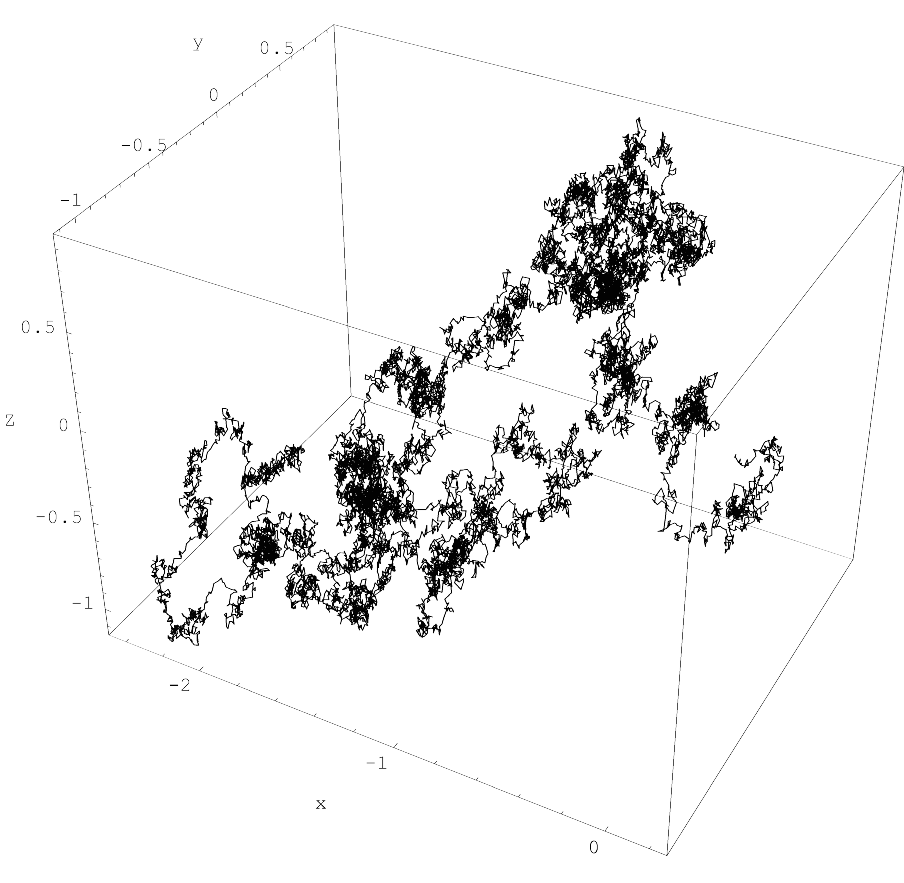
Hyperbolic Algorithm adds second-order derivatives to Langevin dynamics, reducing systematic errors to O(ε²) for lattice …
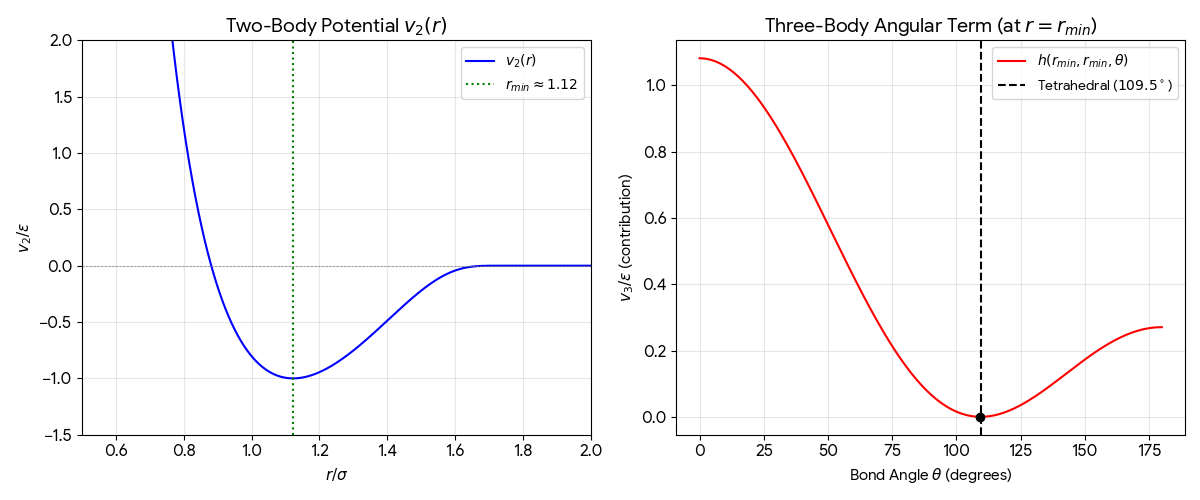
The 1985 paper introducing the Stillinger-Weber potential, a 3-body interaction model for molecular dynamics of …

A 1986 validation of the Evans NEMD method for simulating heat flow, identifying long-time tail anomalies near the …
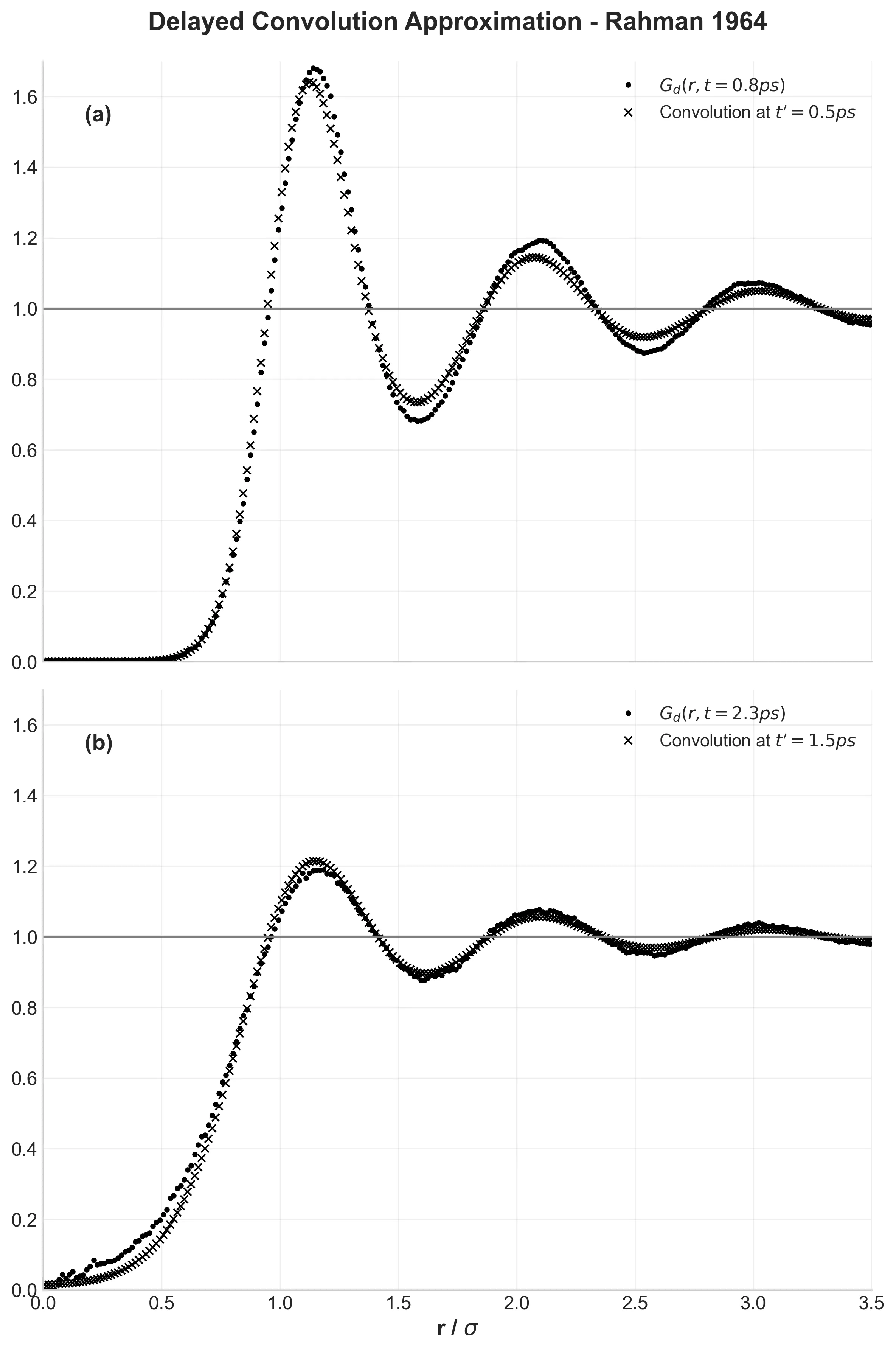
Rahman's foundational 1964 MD simulation of 864 argon atoms revealing the cage effect and validating classical molecular …
A 1984 molecular dynamics study identifying simultaneous multiple jumps in adatom dimer diffusion on fcc(111) surfaces.