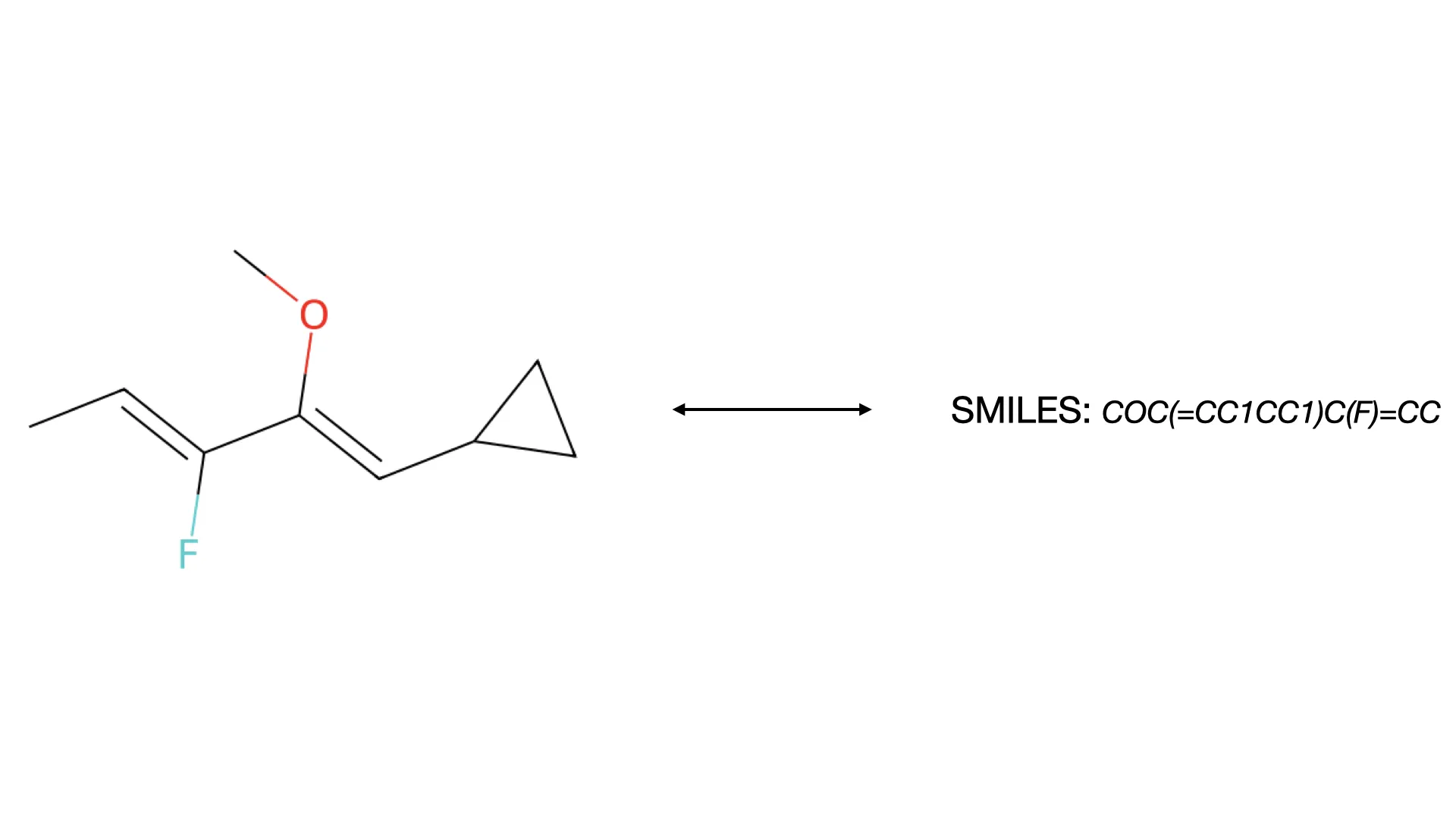
What is Optical Chemical Structure Recognition (OCSR)?
A micro-review of Optical Chemical Structure Recognition (OCSR), covering rule-based systems to modern deep learning …

A micro-review of Optical Chemical Structure Recognition (OCSR), covering rule-based systems to modern deep learning …
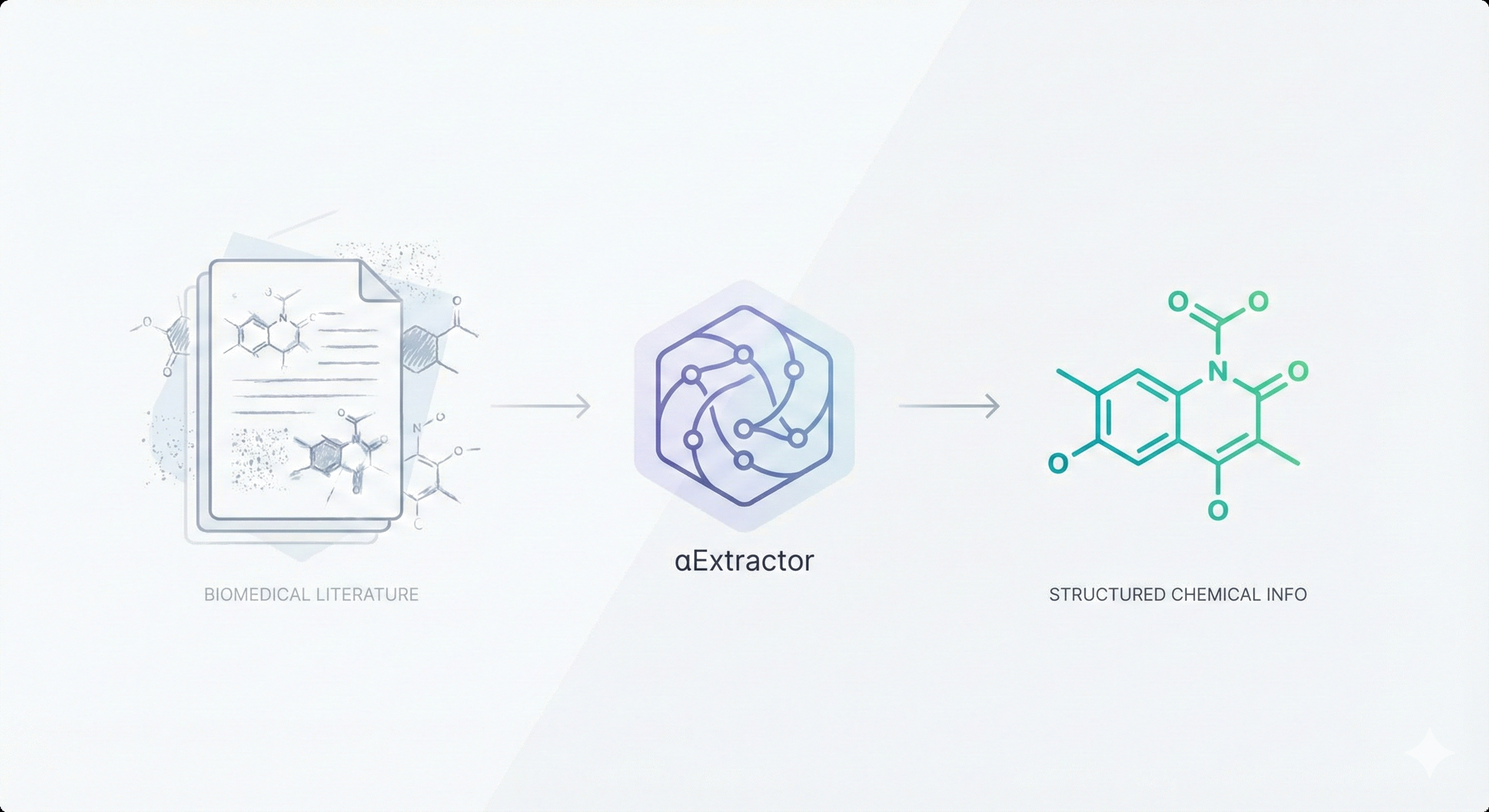
αExtractor uses ResNet-Transformer to extract chemical structures from literature images, including noisy and hand-drawn …
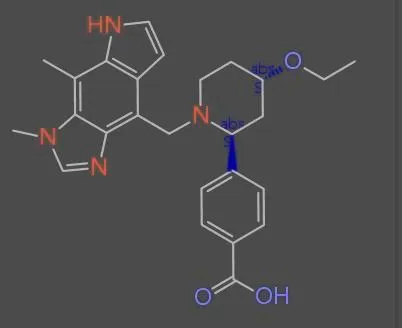
MolParser-7M is the largest OCSR dataset with 7.7M image-text pairs of molecules and E-SMILES, including 400k real-world …
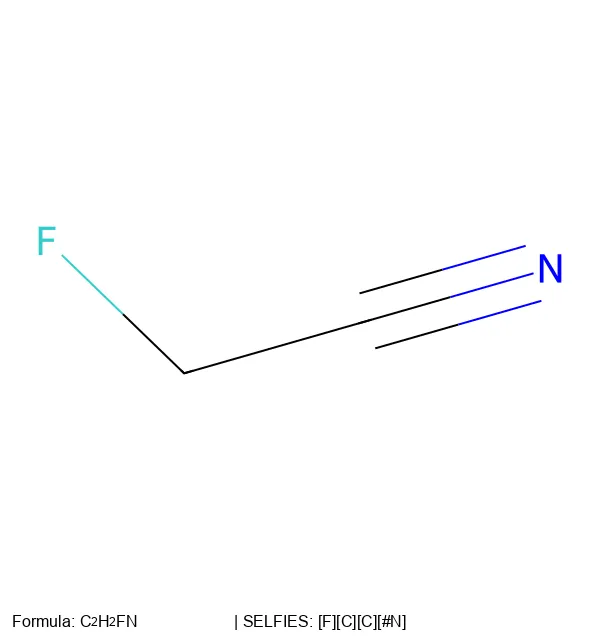
Create 2D molecular images from SELFIES strings using RDKit, SELFIES, and PIL, with proper formatting and legends.
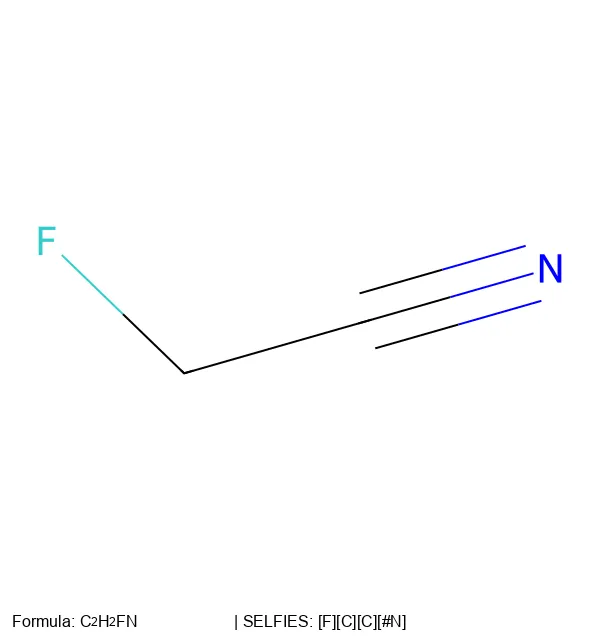
SELFIES is a 100% robust molecular string representation for ML, implemented in the open-source selfies Python library.

Guide to implementing the Müller-Brown potential in PyTorch, comparing analytical vs automatic differentiation with …
Liu et al.'s ICLR 2025 paper introducing DenoiseVAE, which learns adaptive, atom-specific noise for better molecular …
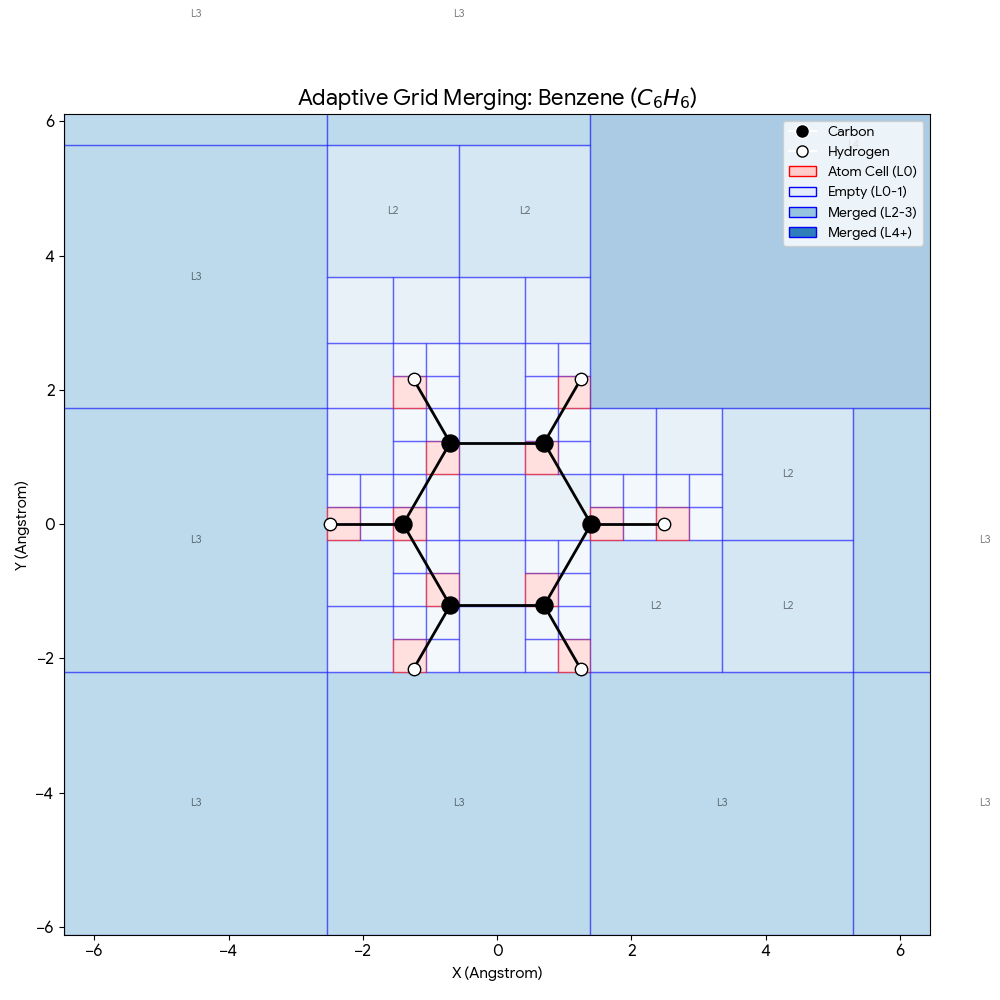
Lu et al. introduce SpaceFormer, a Transformer that models entire 3D molecular space (not just atoms) for superior …
Bigi et al. critique non-conservative force models in ML potentials, showing their simulation failures and proposing …
Luo et al. introduce SPHNet, using adaptive sparsity to dramatically improve SE(3)-equivariant Hamiltonian prediction …

Fu et al. propose energy conservation as a key MLIP diagnostic and introduce eSEN, bridging test accuracy and real …
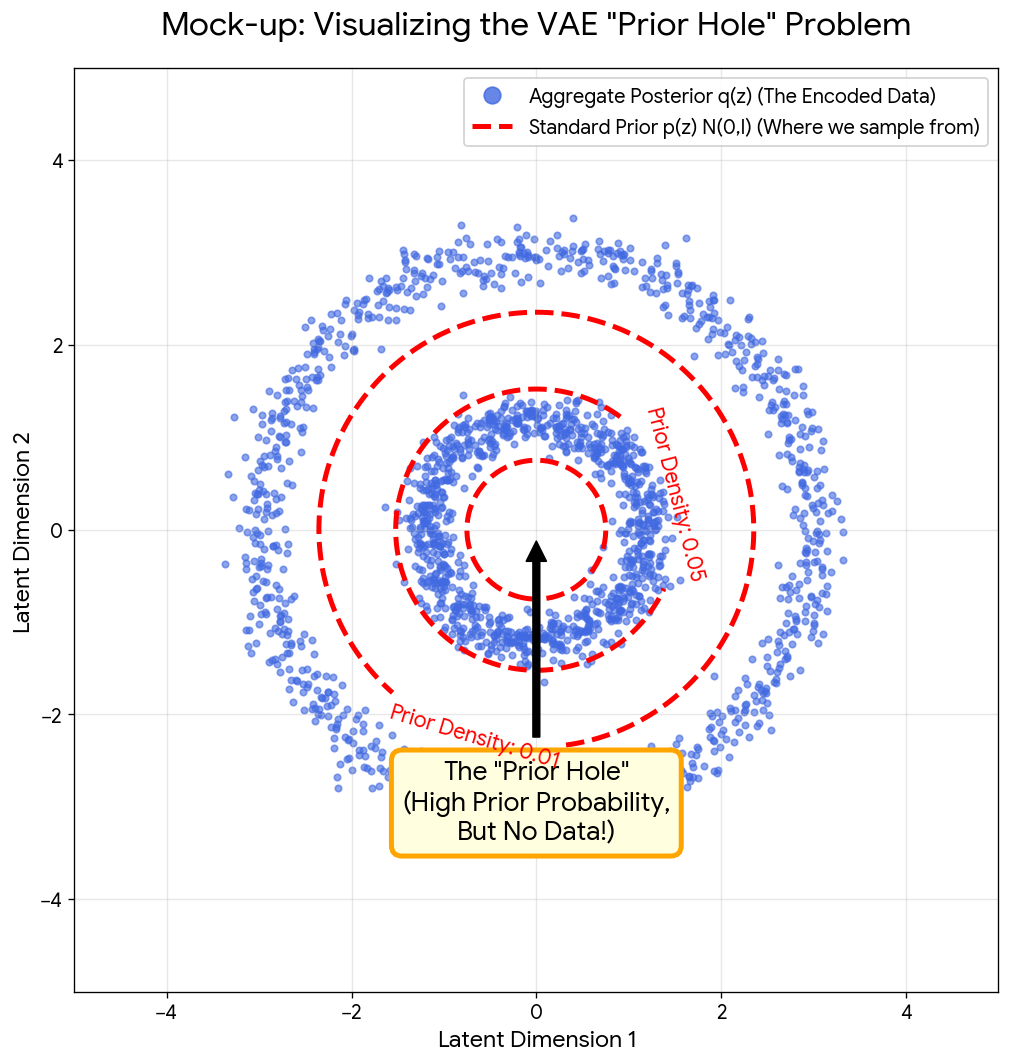
Aneja et al.'s NeurIPS 2021 paper introducing Noise Contrastive Priors (NCPs) to address VAE's 'prior hole' problem with …