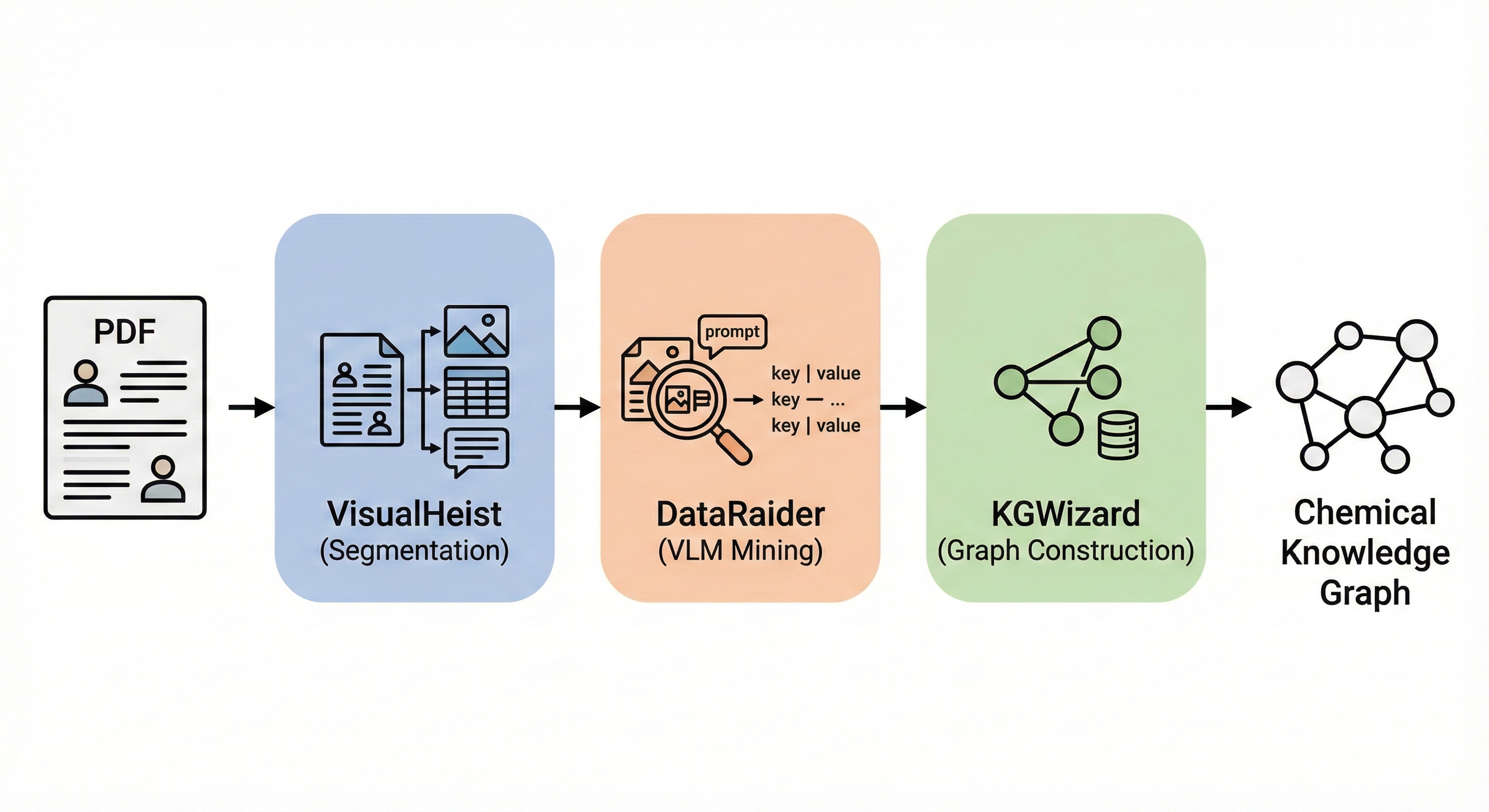
MERMaid: Multimodal Reaction Mining
Vision-language pipeline extracting chemical reaction data from PDF figures and tables into structured knowledge graphs …

Vision-language pipeline extracting chemical reaction data from PDF figures and tables into structured knowledge graphs …
A diffusion-based data augmentation pipeline (OCSAug) using DDPM and RePaint to improve optical chemical structure …
A Transformer-based model for translating SMILES to IUPAC names, trained on ~1 billion molecules, achieving ~99% …
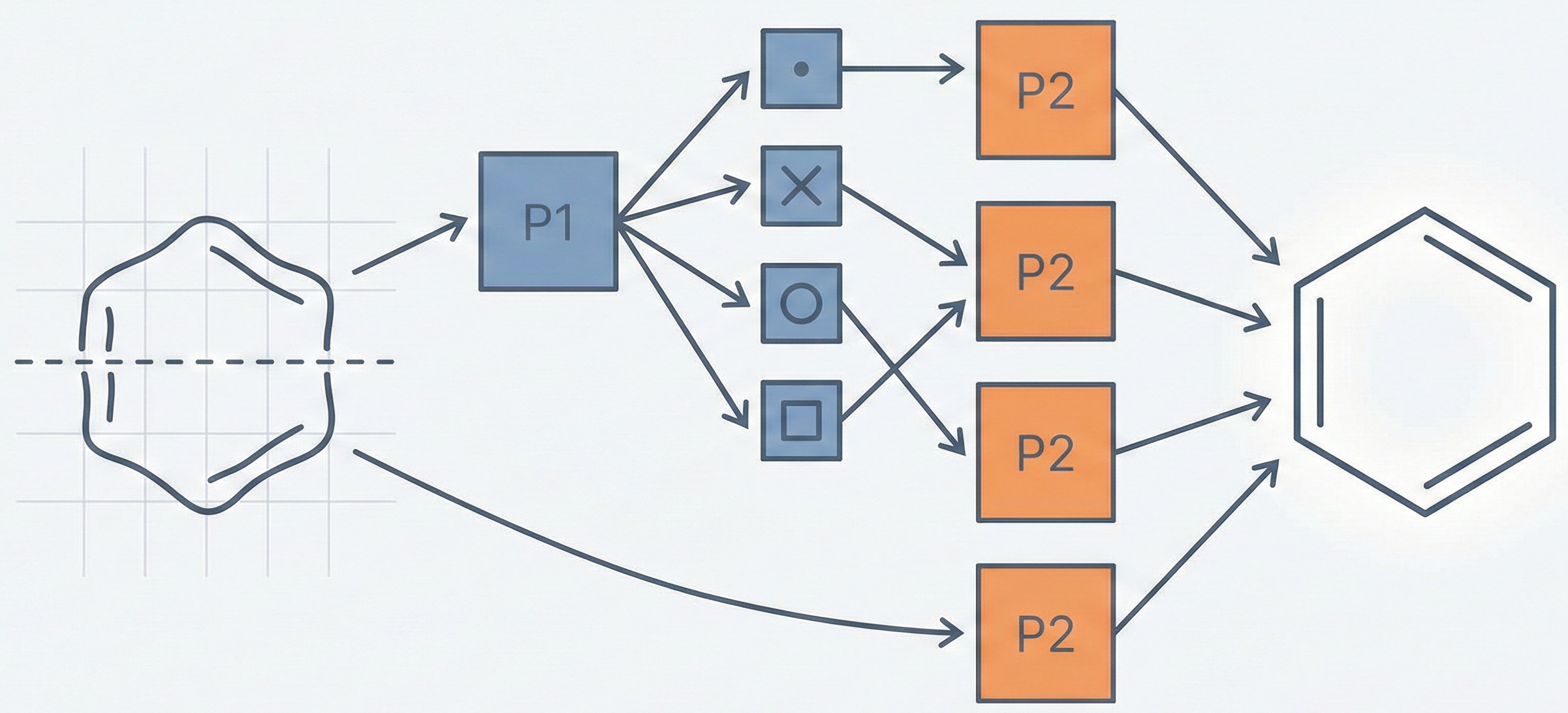
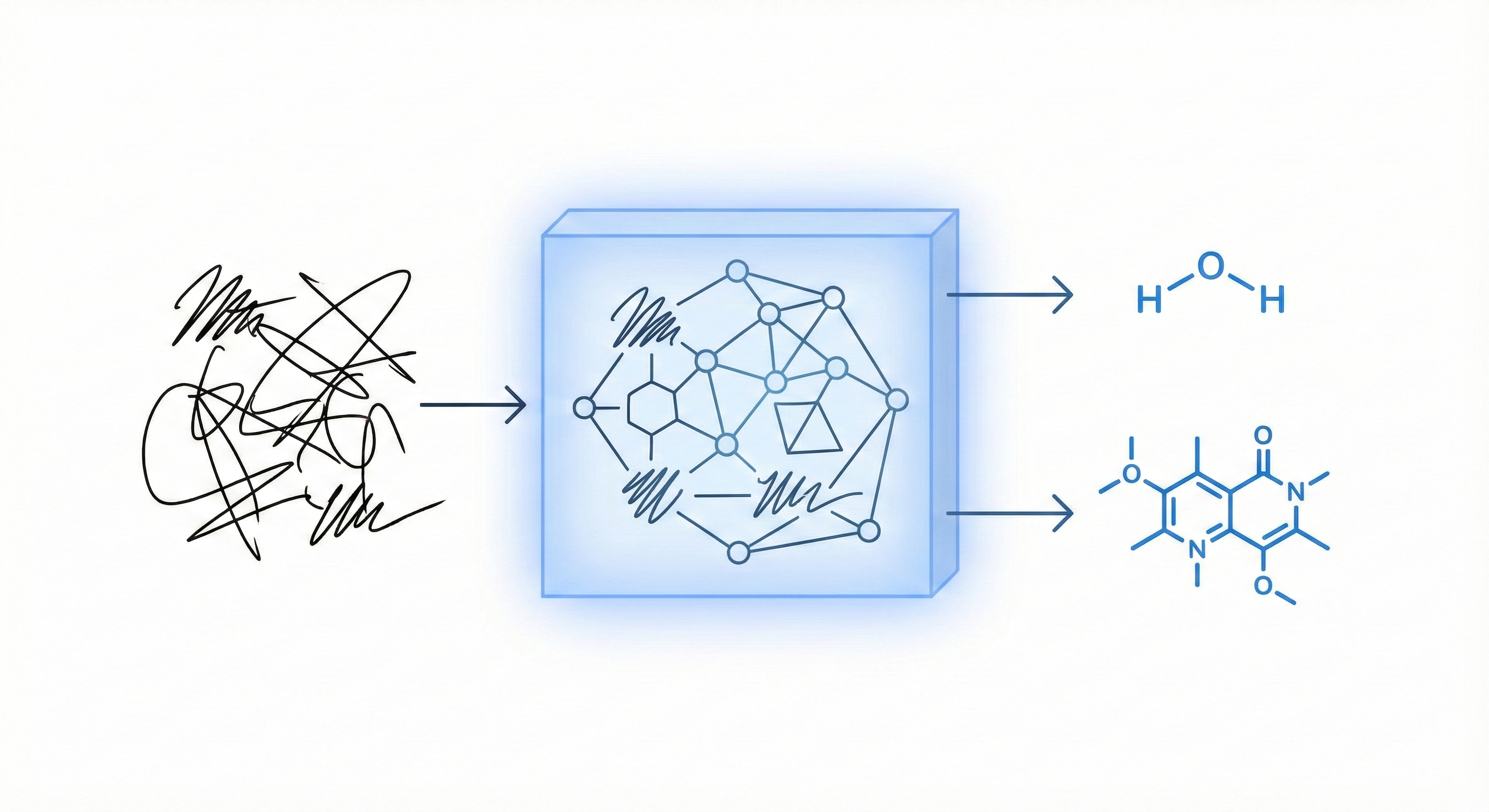
A two-phase neural network approach for recognizing handwritten heterocyclic chemical rings with ~94% accuracy.
A hybrid SVM and elastic matching approach for recognizing handwritten chemical symbols drawn on touch devices, …
Online recognition of handwritten chemical symbols using Hidden Markov Models with 11-dimensional local features, …
Two-level algorithm for recognizing on-line handwritten chemical expressions using structural analysis, ANNs, and string …
A hierarchical grammar-based approach for recognizing and analyzing online handwritten chemical formulas in mobile …
A two-level recognition algorithm for on-line handwritten chemical expressions using structural and syntactic features.
A dual-stage classifier combining SVM and HMM to recognize online handwritten chemical symbols, introducing a reordering …
A 2009 unified framework for inorganic/organic chemical handwriting recognition using graph search and statistical …
A probabilistic approach using Markov Logic Networks to recognize chemical structures from images, improving robustness …