
GutenOCR: A Grounded Vision-Language Front-End for Documents
GutenOCR is a family of vision-language models designed to serve as a ‘grounded OCR front-end’, providing high-quality text transcription and explicit geometric grounding.

GutenOCR is a family of vision-language models designed to serve as a ‘grounded OCR front-end’, providing high-quality text transcription and explicit geometric grounding.
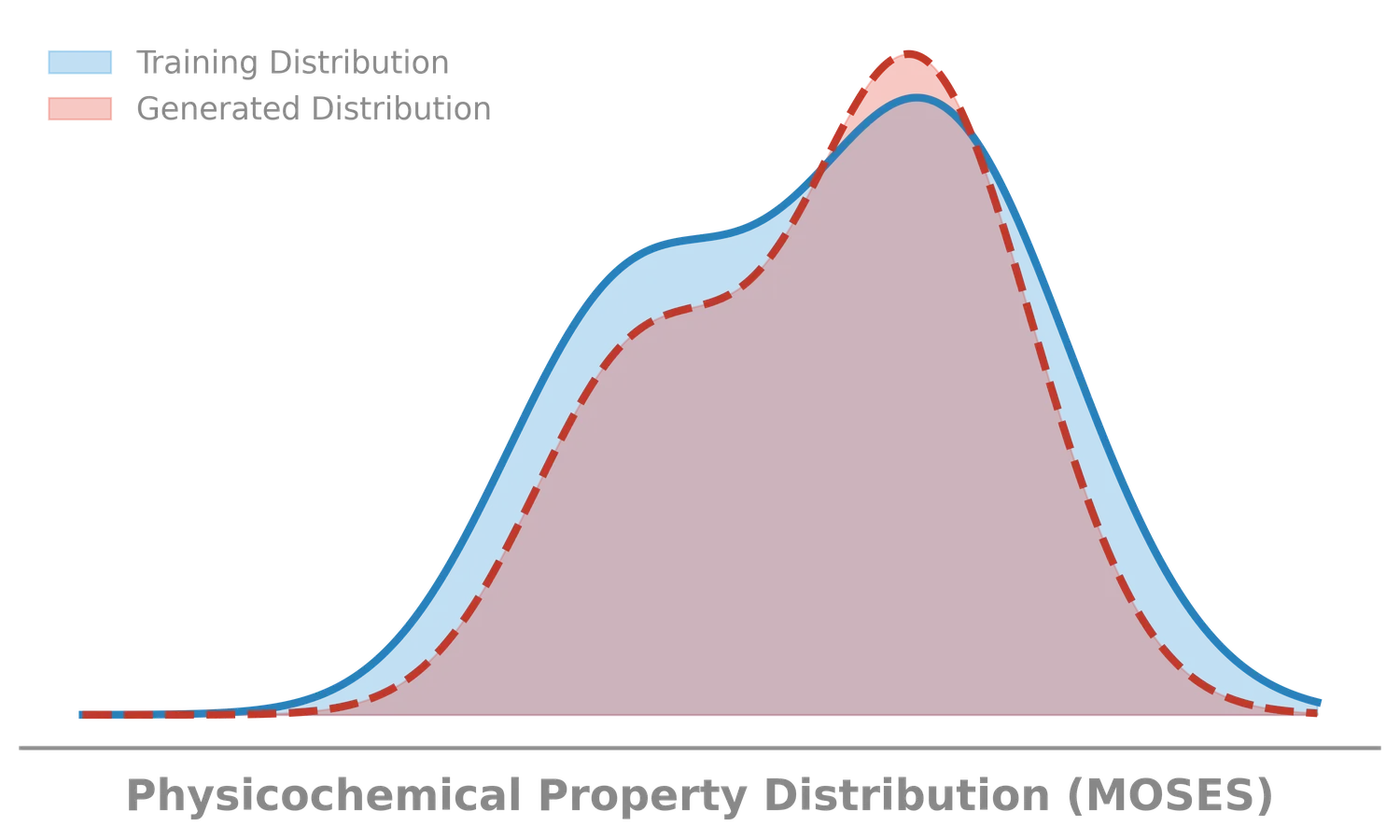
MOSES introduces a comprehensive benchmarking platform for molecular generative models, offering standardized datasets, evaluation metrics, and baselines. By providing a unified measuring stick, it aims to resolve reproducibility challenges in chemical distribution learning.
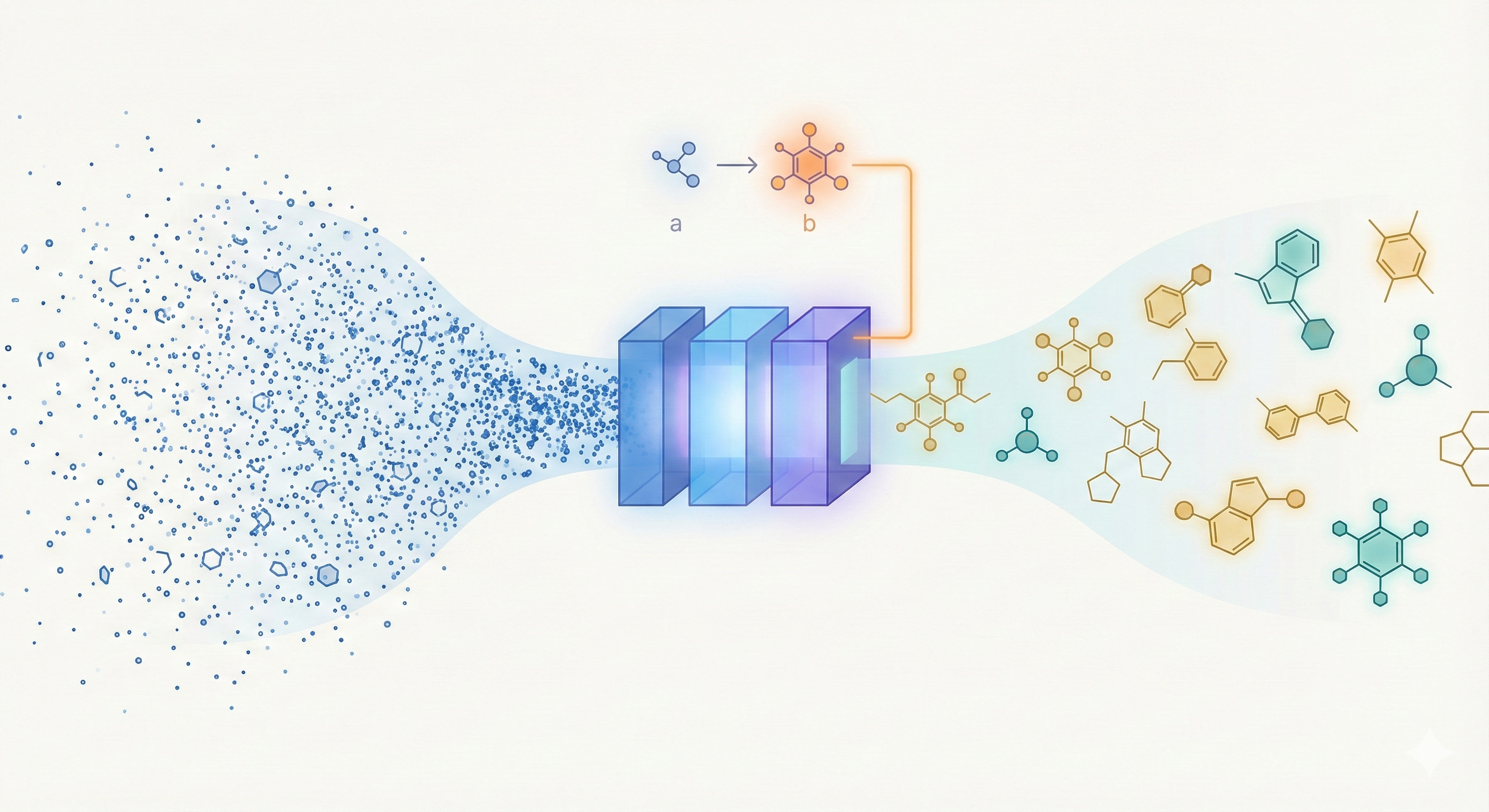
This methodological paper proposes a linear-attention transformer decoder trained on 1.1 billion molecules. It introduces pair-tuning for efficient property optimization and establishes empirical scaling laws relating inference compute to generation novelty.
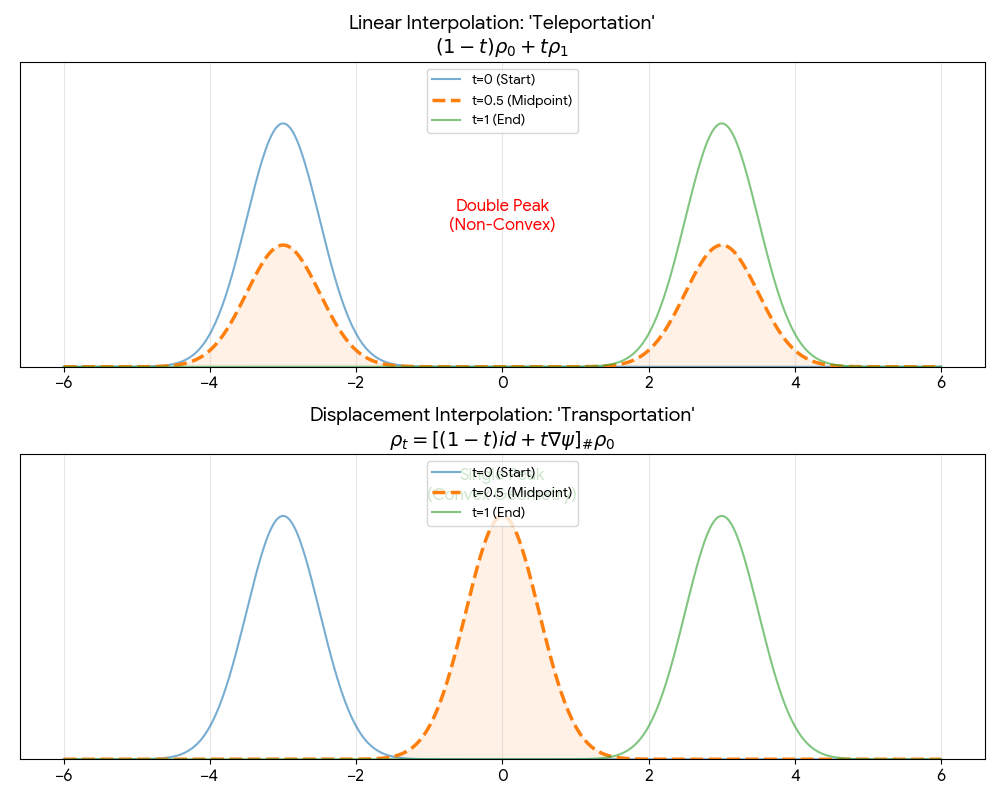
A foundational theoretical paper that introduces displacement interpolation (optimal transport) to establish a new convexity principle for energy functionals. It proves the uniqueness of ground states for interacting gases and generalizes the Brunn-Minkowski inequality, providing mathematical foundations used in modern generative models.
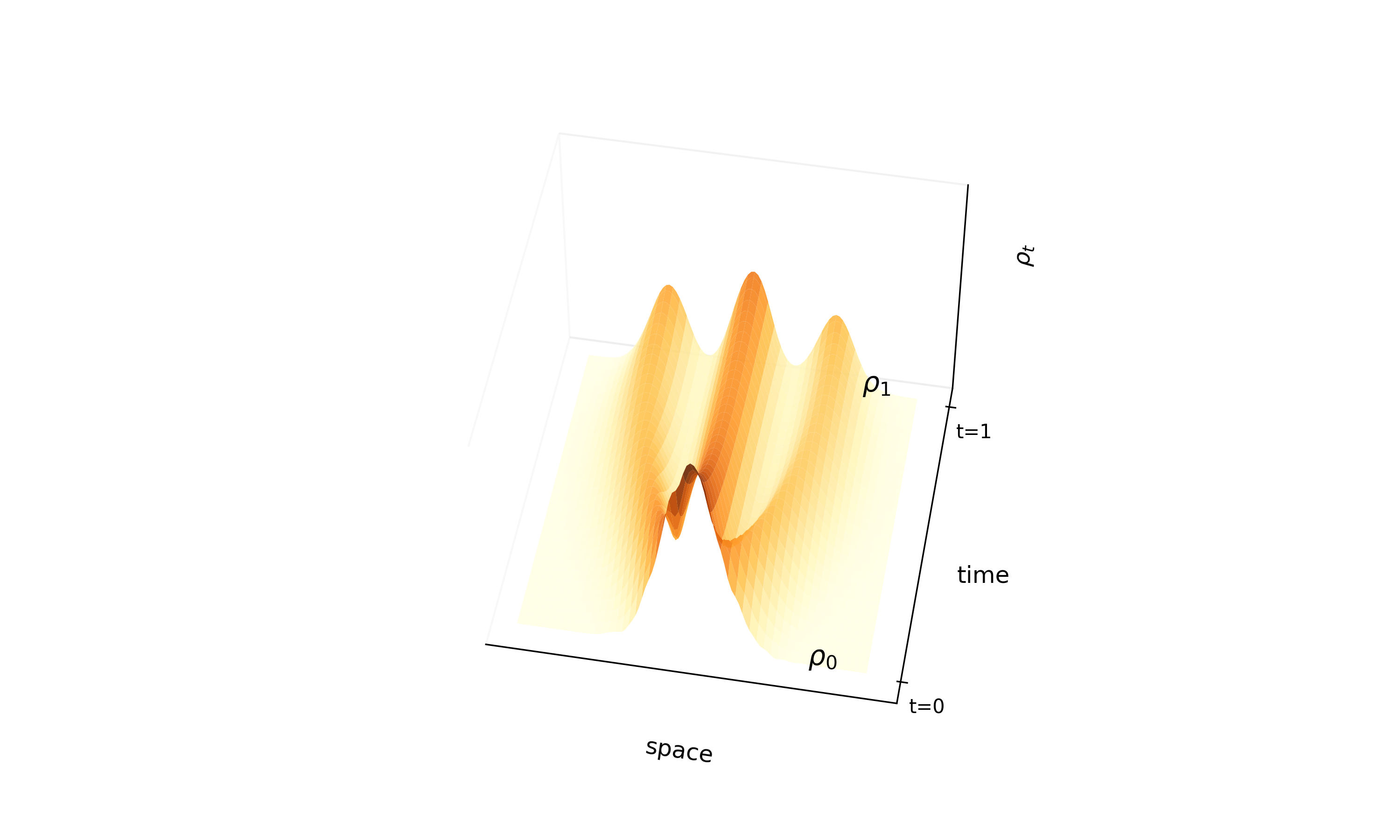
Proposes ‘InterFlow’, a method to learn continuous normalizing flows between arbitrary densities using stochastic interpolants. It avoids ODE backpropagation by minimizing a quadratic objective on the velocity field, enabling scalable ODE-based generation.
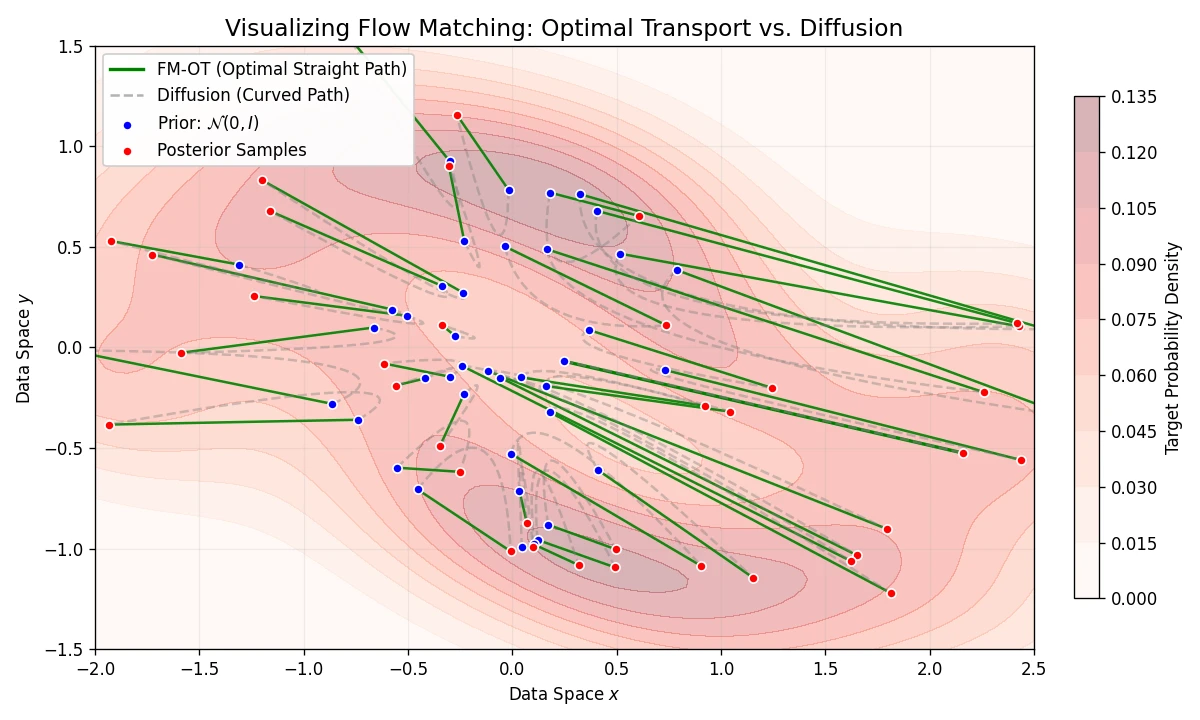
Introduces Flow Matching, a scalable method for training CNFs by regressing vector fields of conditional probability paths. It generalizes diffusion and enables Optimal Transport paths for straighter, more efficient sampling.
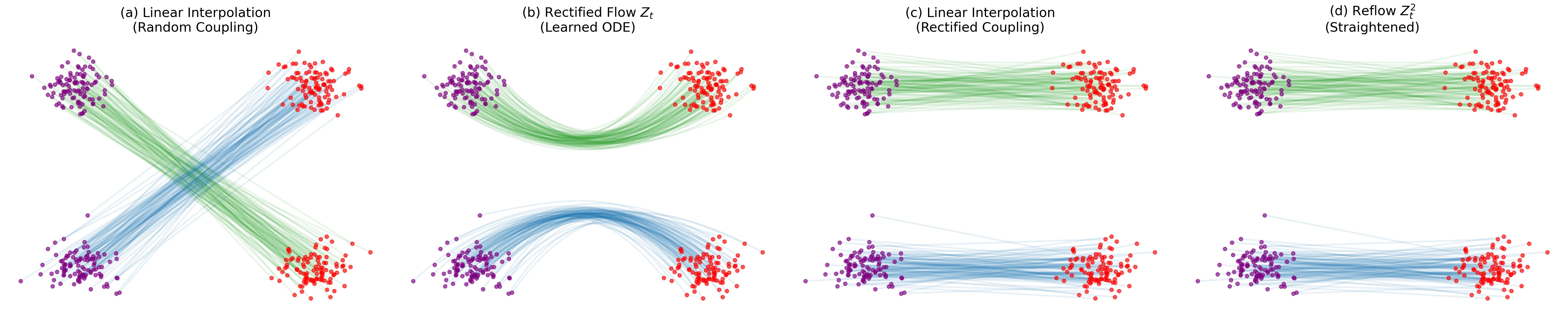
Introduces ‘Rectified Flow,’ a method to transport distributions via ODEs with straight paths. Uses a ‘reflow’ procedure to iteratively straighten trajectories, enabling high-quality 1-step generation without complex distillation pipelines.
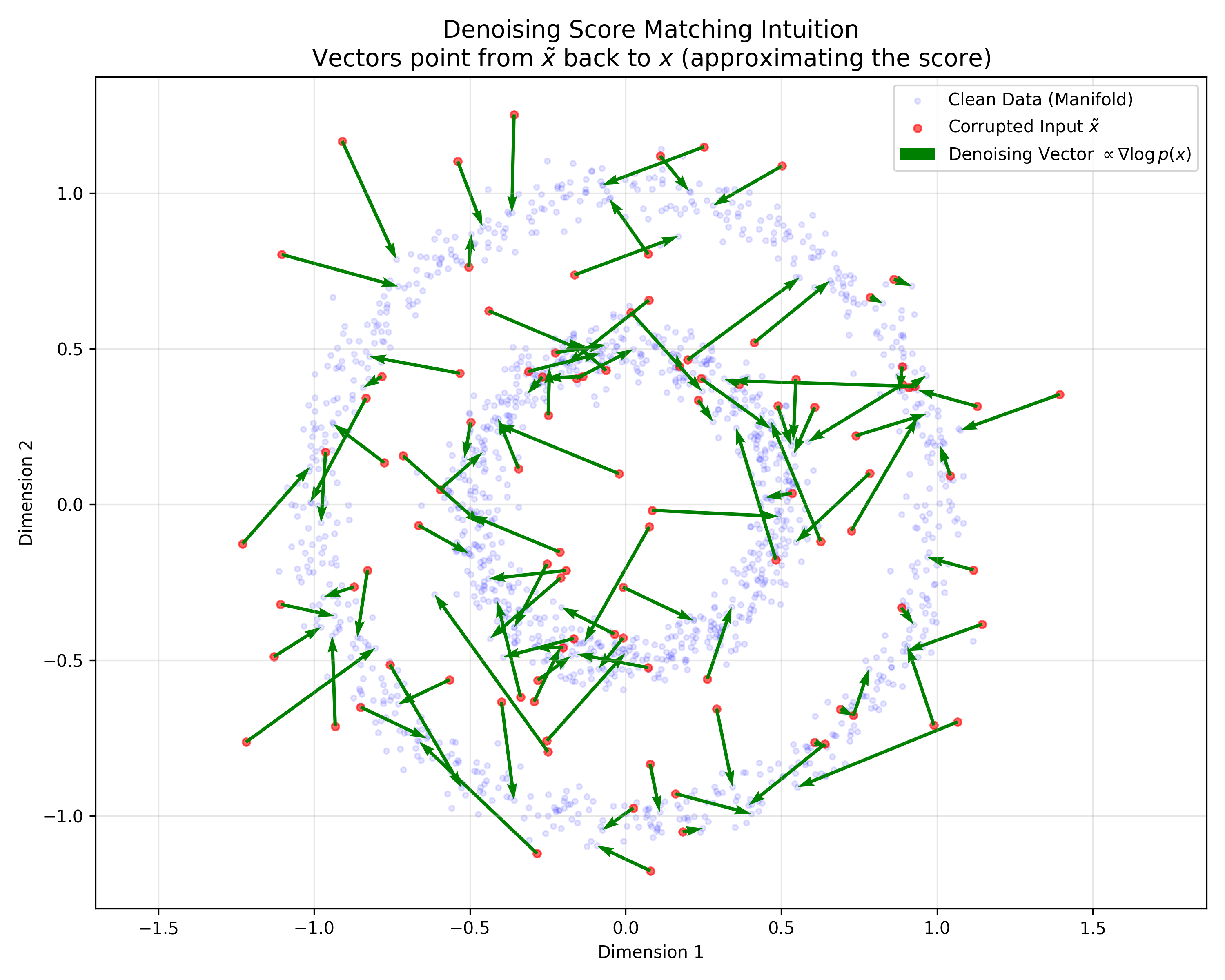
This paper provides a rigorous probabilistic foundation for Denoising Autoencoders by proving they are mathematically equivalent to Score Matching on a kernel-smoothed data distribution. It derives a specific energy function for DAEs and justifies the use of tied weights.
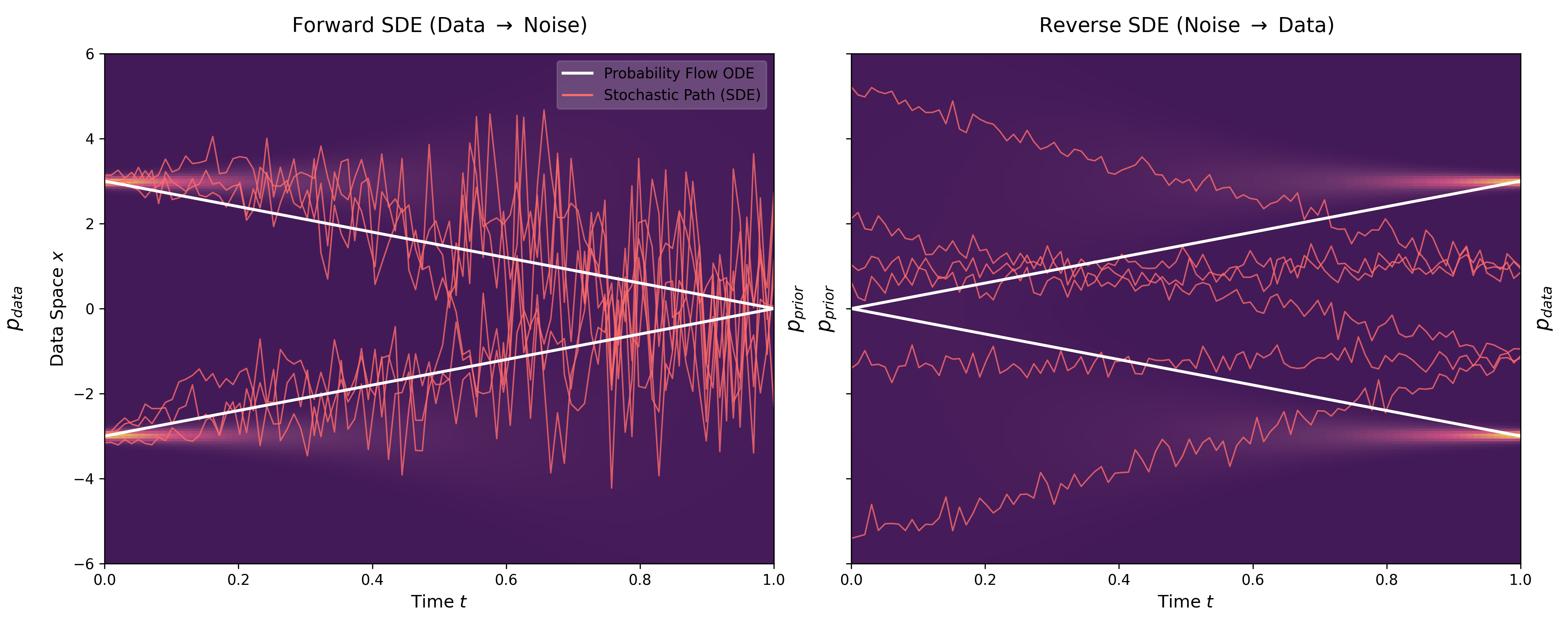
This paper unifies previous score-based methods (SMLD and DDPM) under a continuous-time SDE framework. It introduces Predictor-Corrector samplers for improved generation and Probability Flow ODEs for exact likelihood computation, setting new records on CIFAR-10.
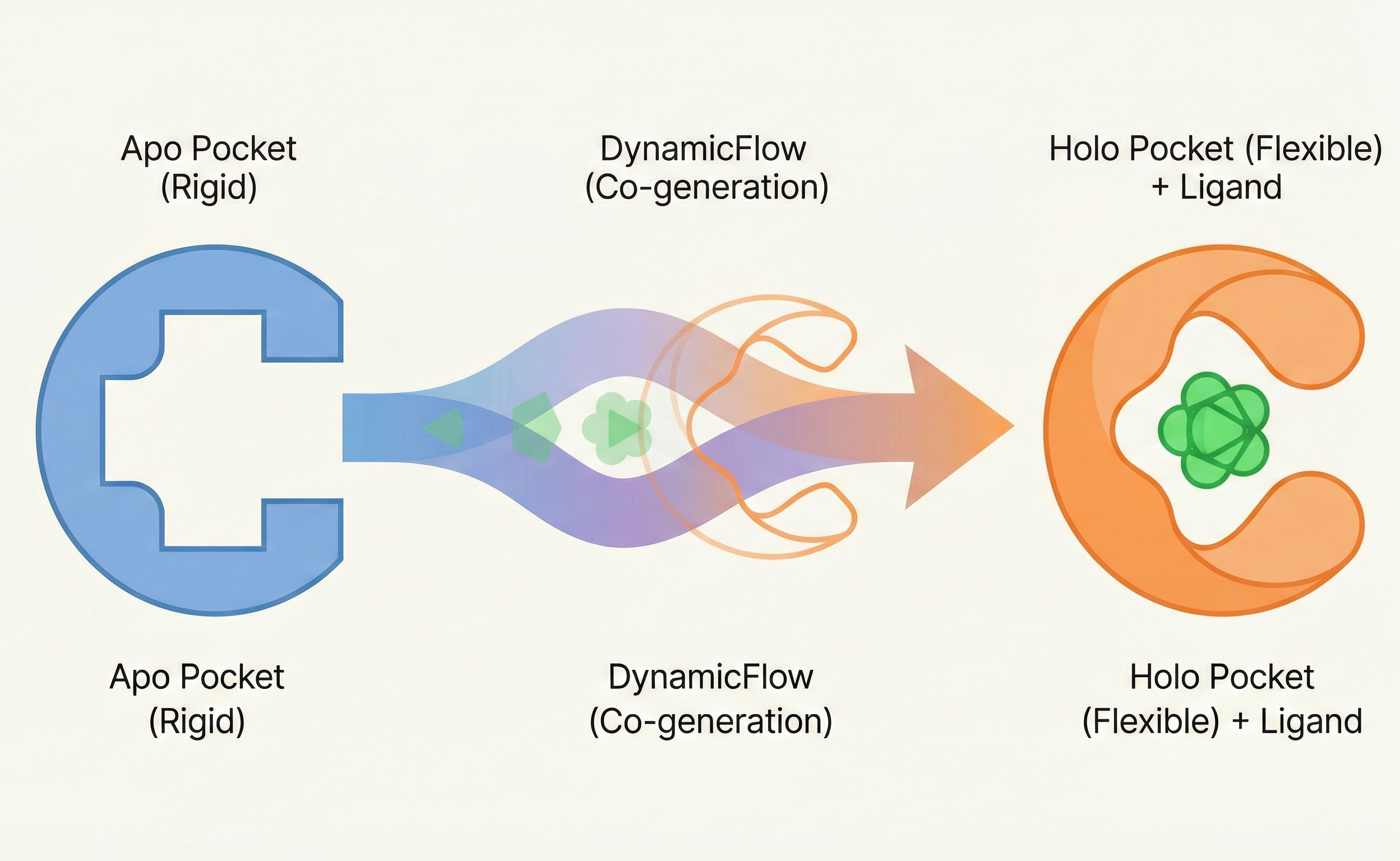
This paper introduces DynamicFlow, a full-atom stochastic flow matching model that simultaneously generates ligand molecules and transforms protein pockets from apo to holo states. It also contributes a new dataset of MD-simulated apo-holo pairs derived from MISATO.
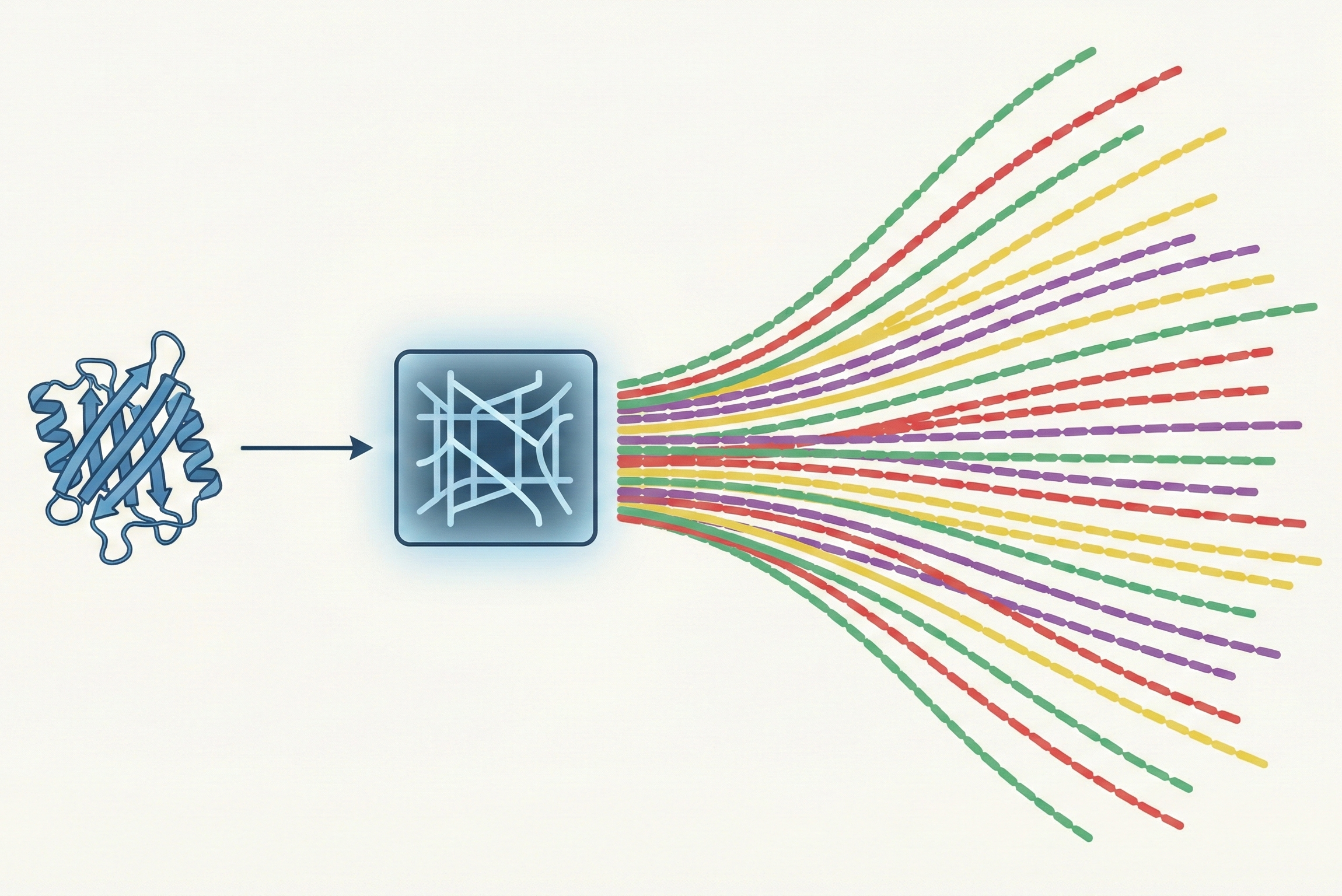
InvMSAFold replaces autoregressive decoding with a Potts model parameter generator, enabling diverse protein sequence sampling orders of magnitude faster than ESM-IF1.
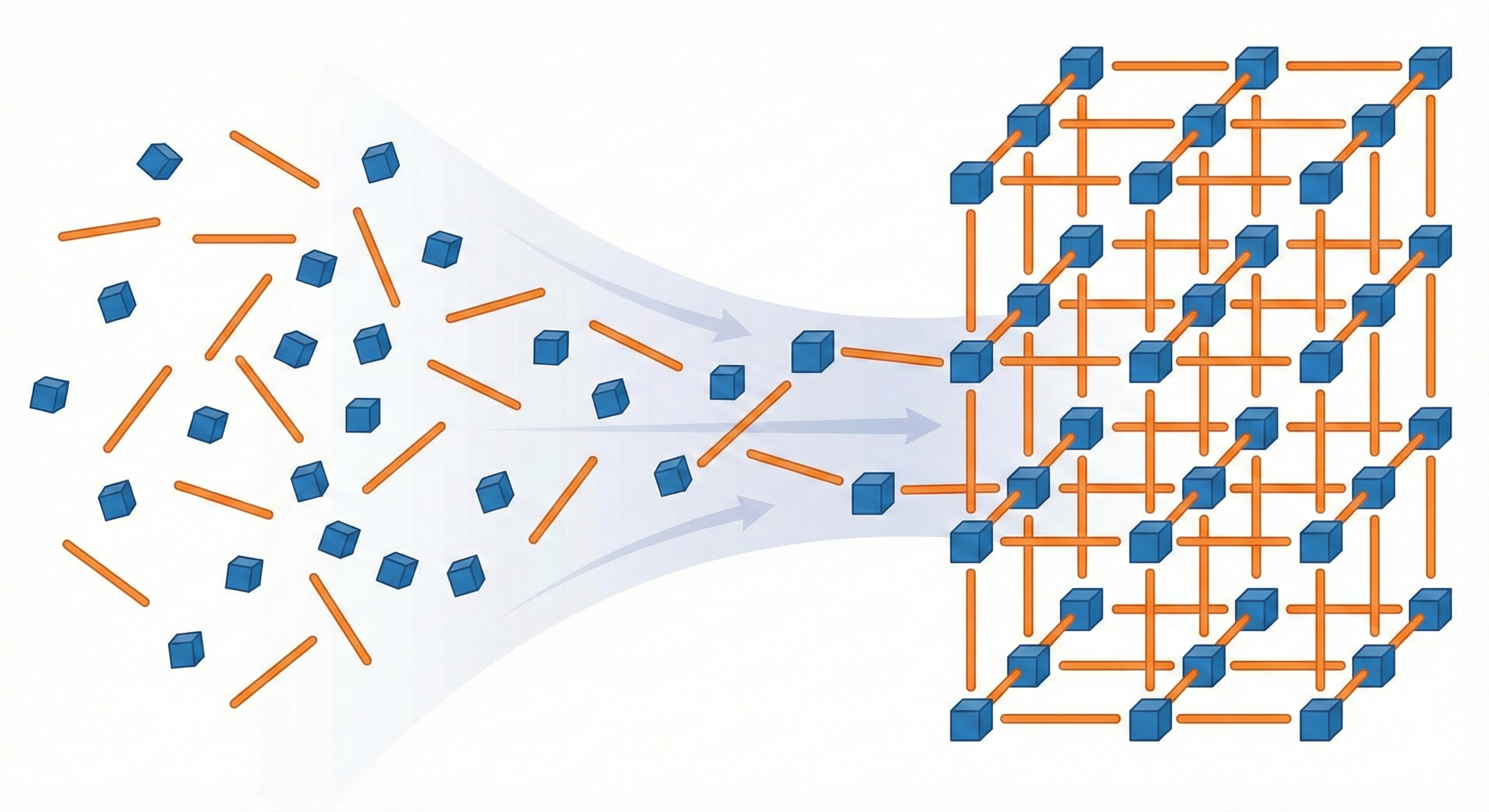
MOFFlow is the first deep generative model tailored for Metal-Organic Framework (MOF) structure prediction. It utilizes Riemannian flow matching on SE(3) to assemble rigid building blocks (metal nodes and organic linkers), achieving significantly higher accuracy and scalability than atom-based methods on large systems.