MARCEL: Molecular Representation and Conformer Ensemble Learning
MARCEL dataset provides 722K+ conformers across 76K+ molecules for drug discovery, catalysis, and molecular …

MARCEL dataset provides 722K+ conformers across 76K+ molecules for drug discovery, catalysis, and molecular …
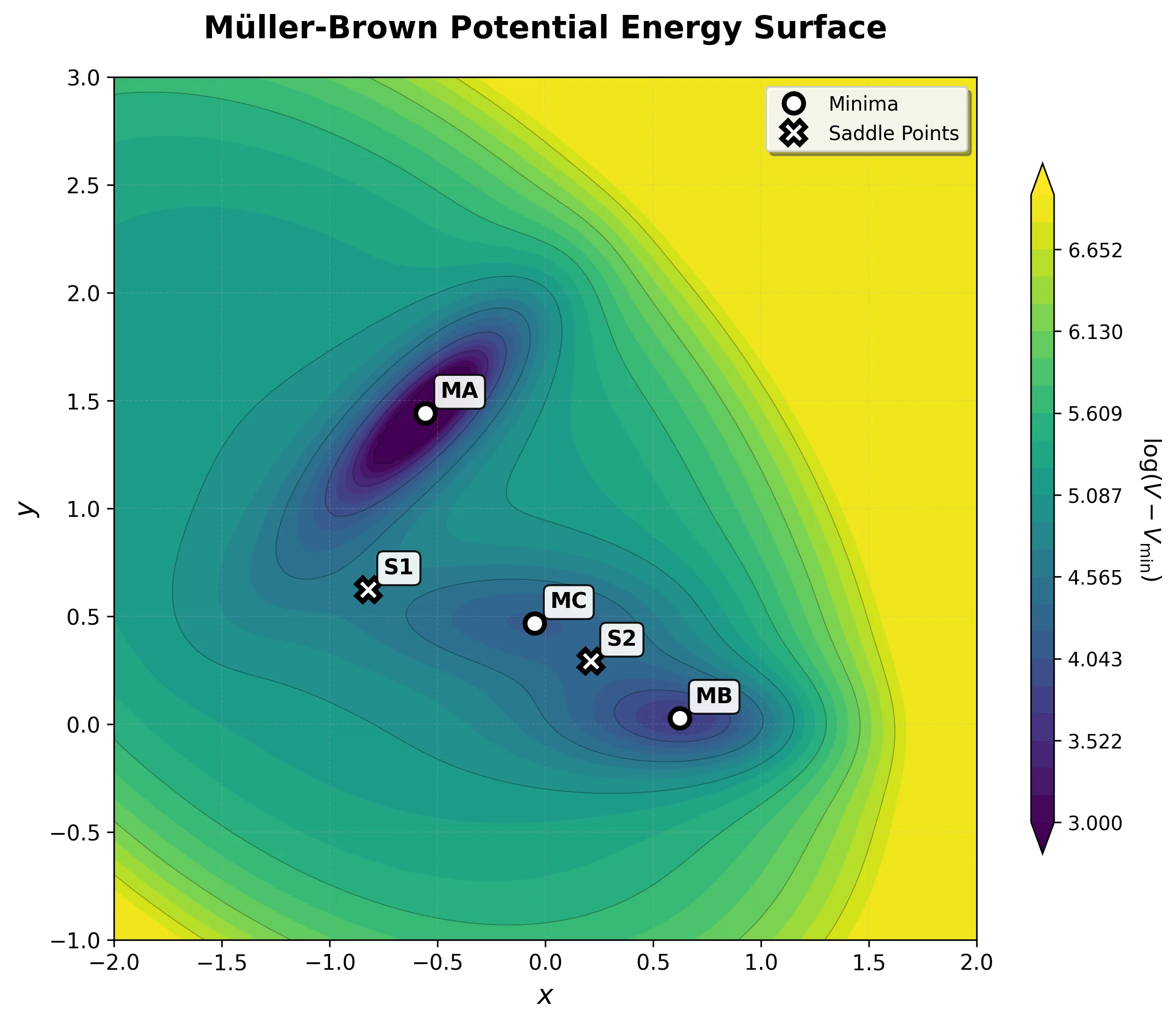
The Müller-Brown potential: a classic two-dimensional analytical benchmark for testing optimization algorithms, reaction …...
SMILES is a specification for describing the structure of chemical molecules using short ASCII strings....
Henze and Blair's 1931 JACS paper introducing the recursive method for counting alkane isomers, founding mathematical …...
A dataset card for the GEOM dataset, a collection of energy-annotated molecular conformations for property prediction …
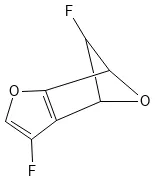
A dataset card for the Generated Database 11 (GDB-11), a database of 26.4 million small organic molecules for virtual …

Guide to implementing the Müller-Brown potential in PyTorch, comparing analytical vs automatic differentiation with …
Langevin dynamics simulation showing particle motion in the deep reactant minimum (Basin MA) of the Müller-Brown …
Langevin dynamics simulation showing particle motion in the product minimum (Basin MB) of the Müller-Brown potential …

PyTorch implementation of the Müller-Brown potential with performance optimizations, MD simulations, and analytical vs …...
Extended Langevin dynamics simulation showing particle transitions between different basins of the Müller-Brown …
Liu et al.'s ICLR 2025 paper introducing DenoiseVAE, which learns adaptive, atom-specific noise for better molecular …...