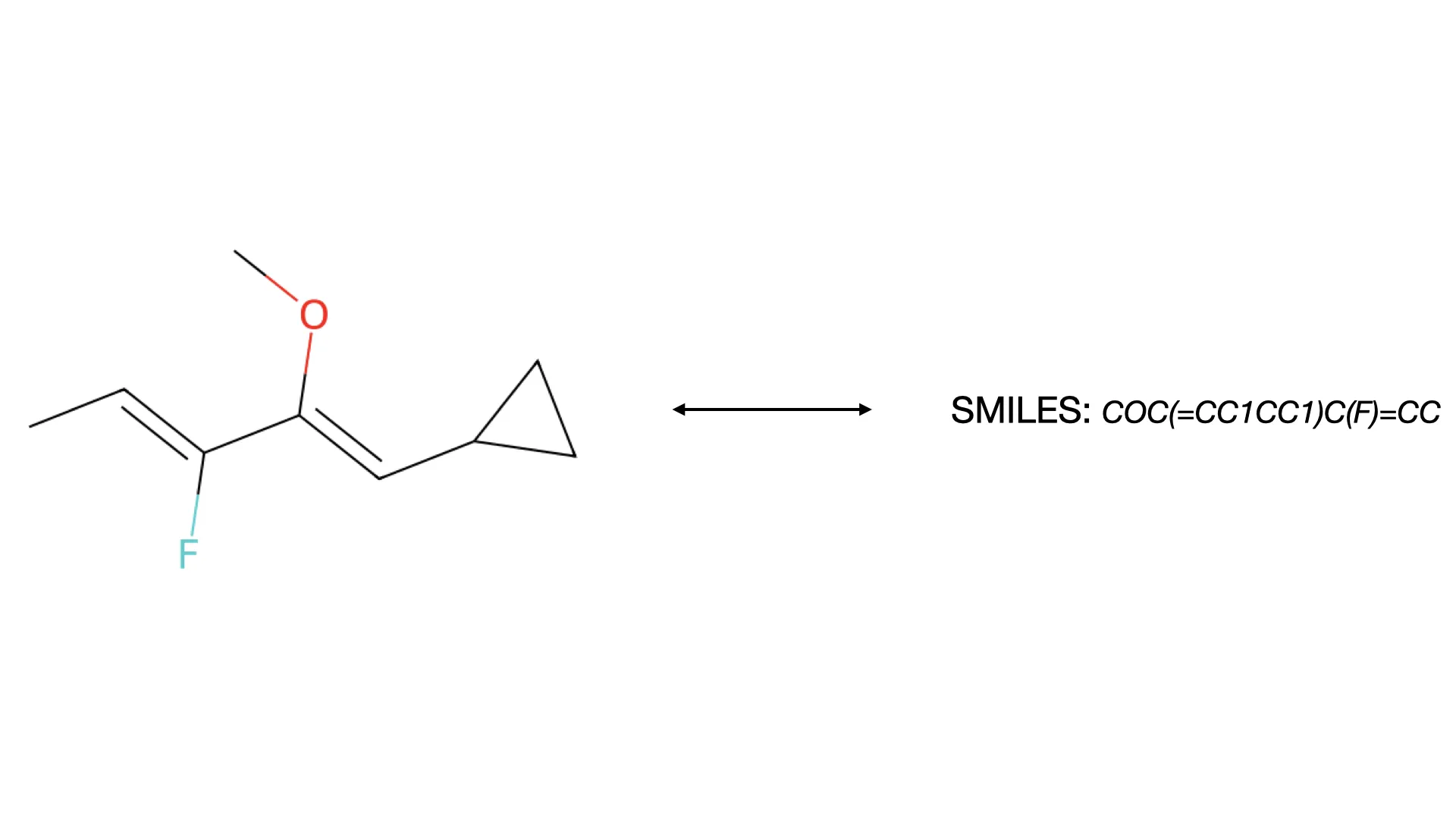
MolParser: End-to-End Molecular Structure Recognition
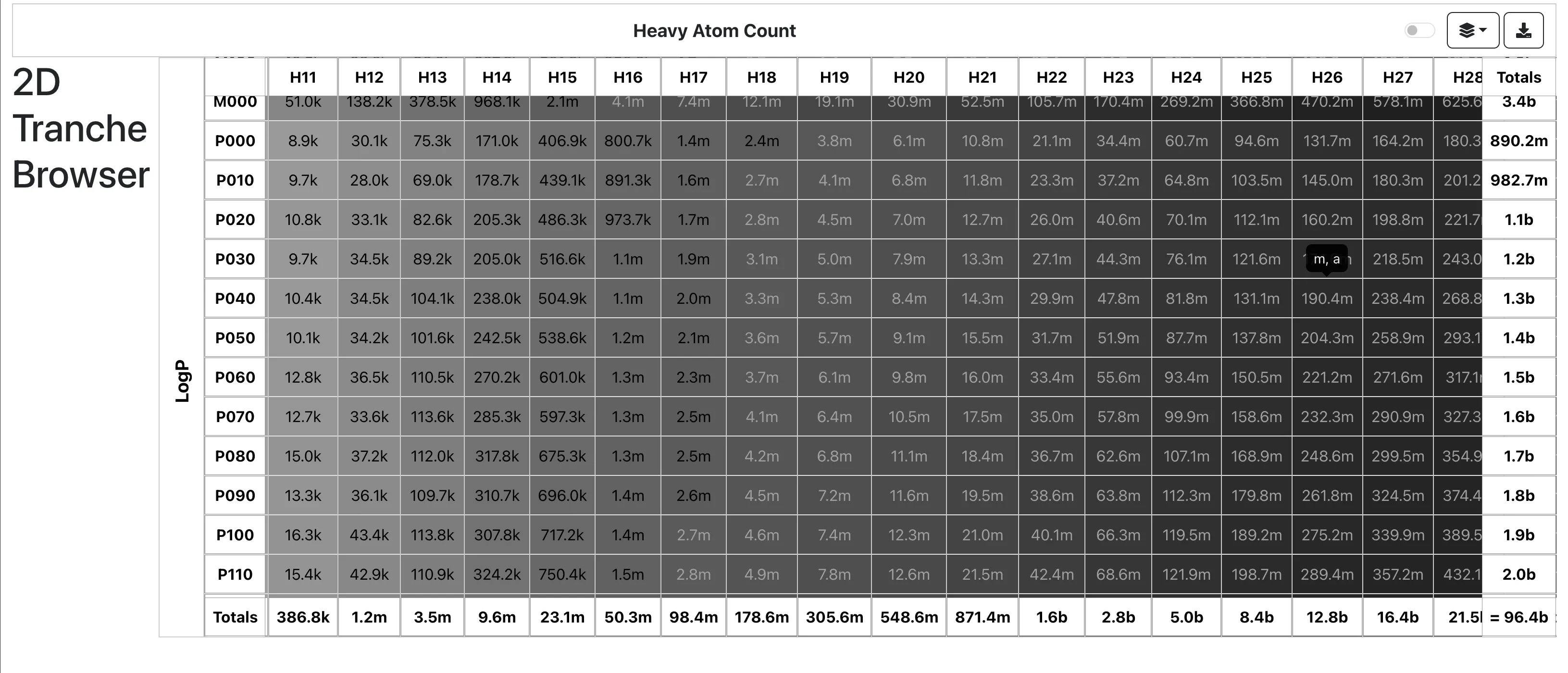
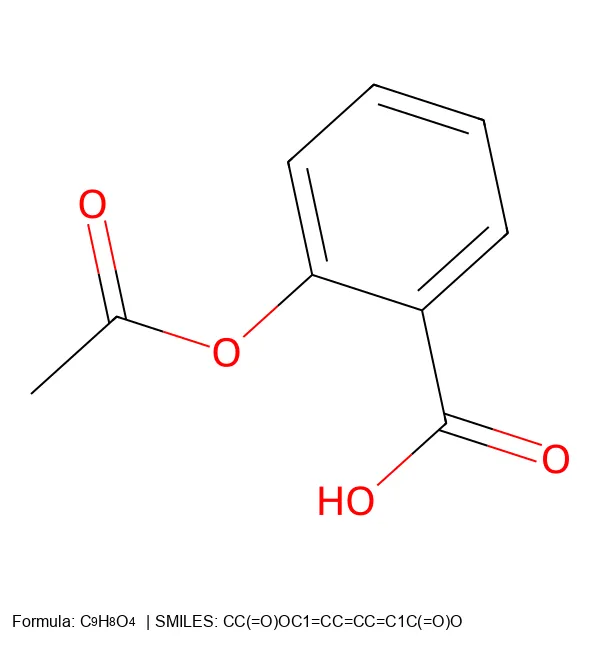
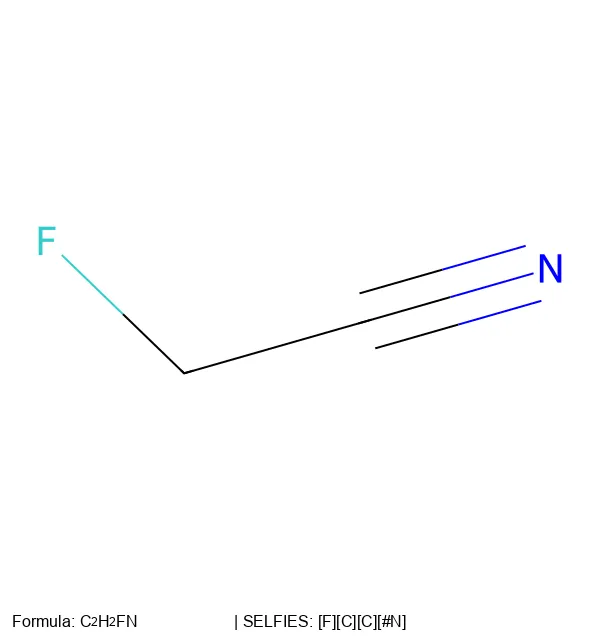
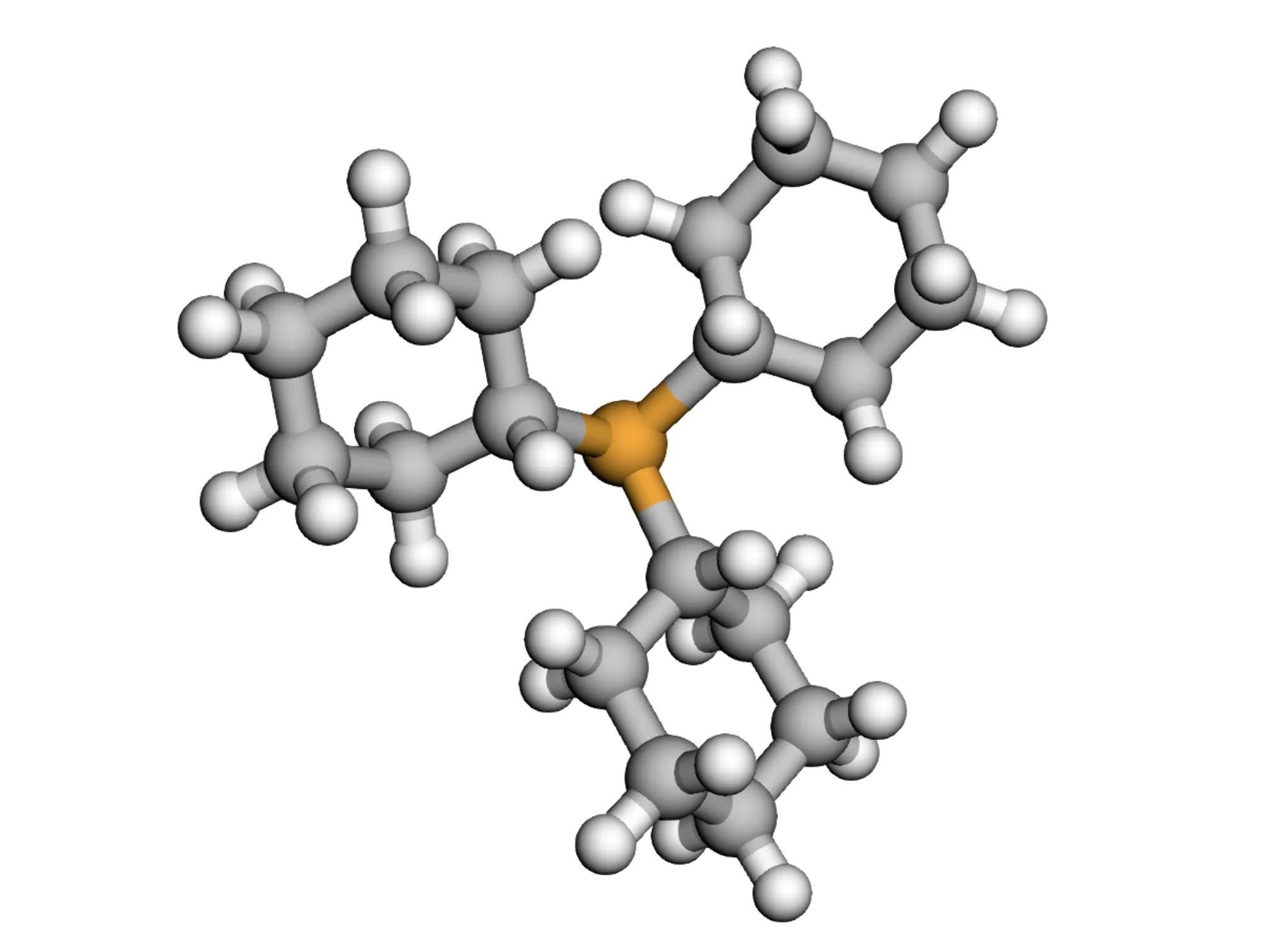
A 2025 end-to-end OCSR system addressing both technical and data challenges, introducing MolParser-7M (7M+ image-text pairs) and MolDet (YOLO-based detector) for extracting and recognizing molecular structures from real-world documents with diverse quality and styles.