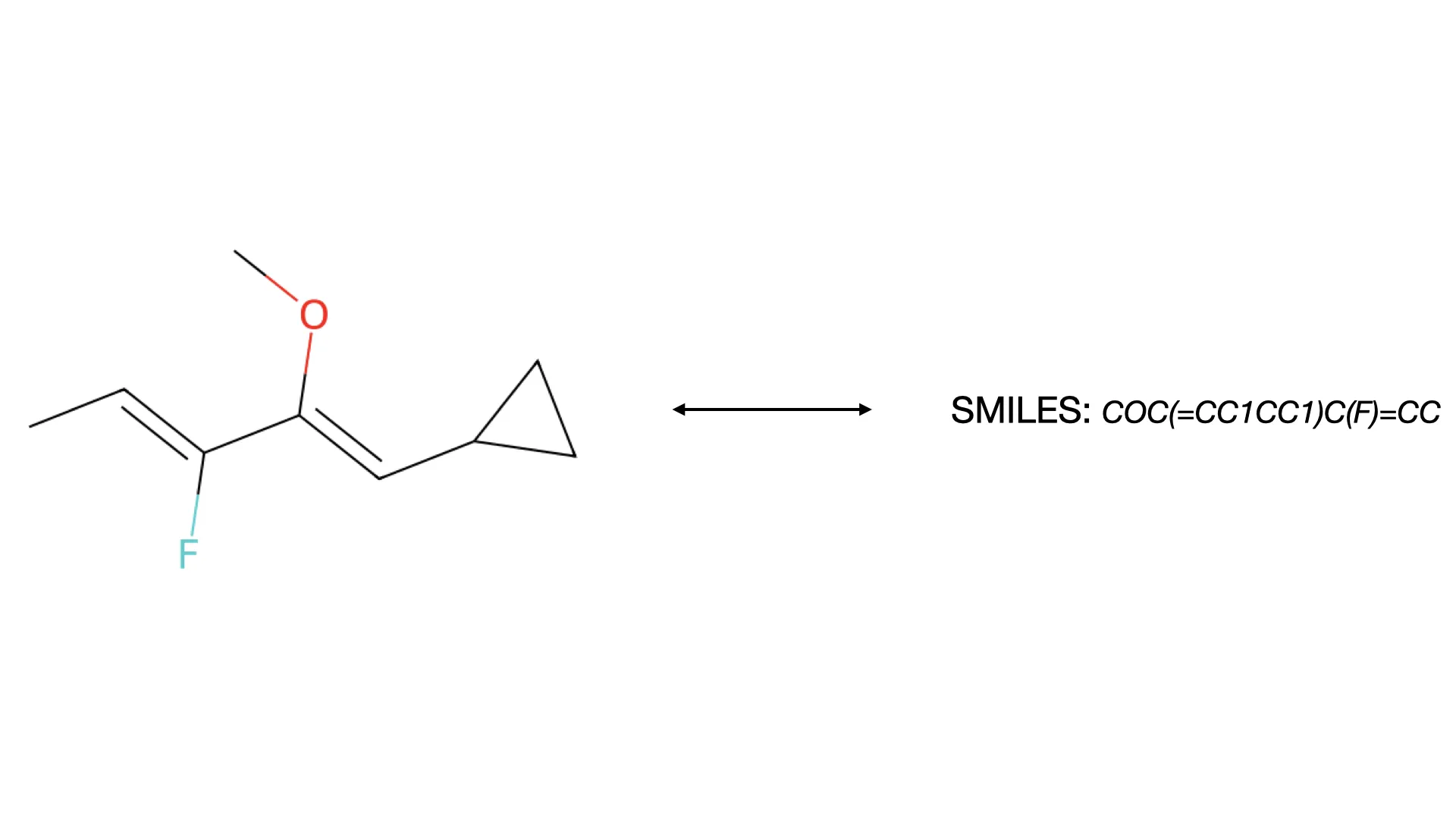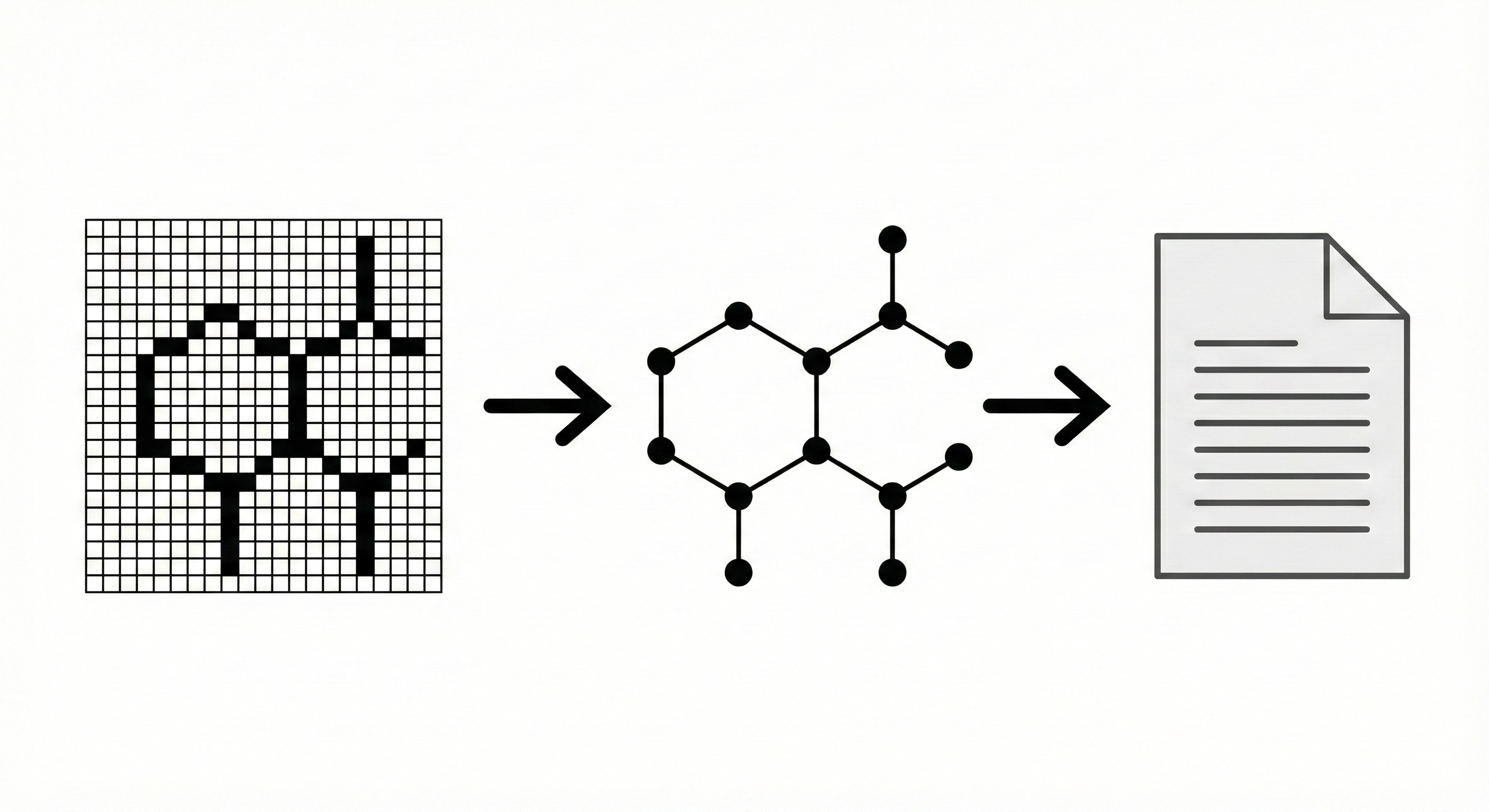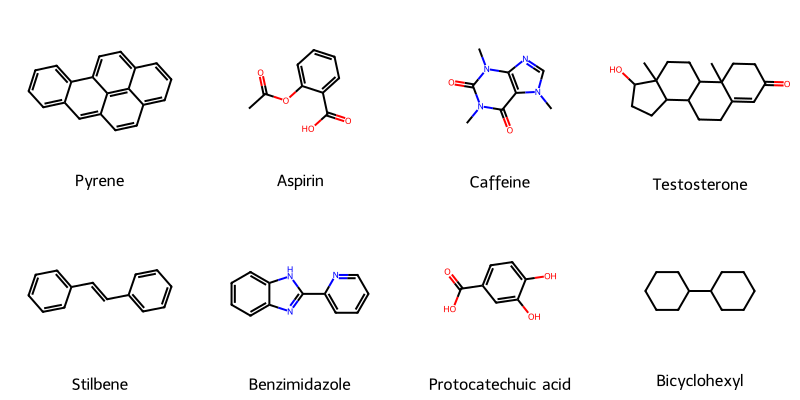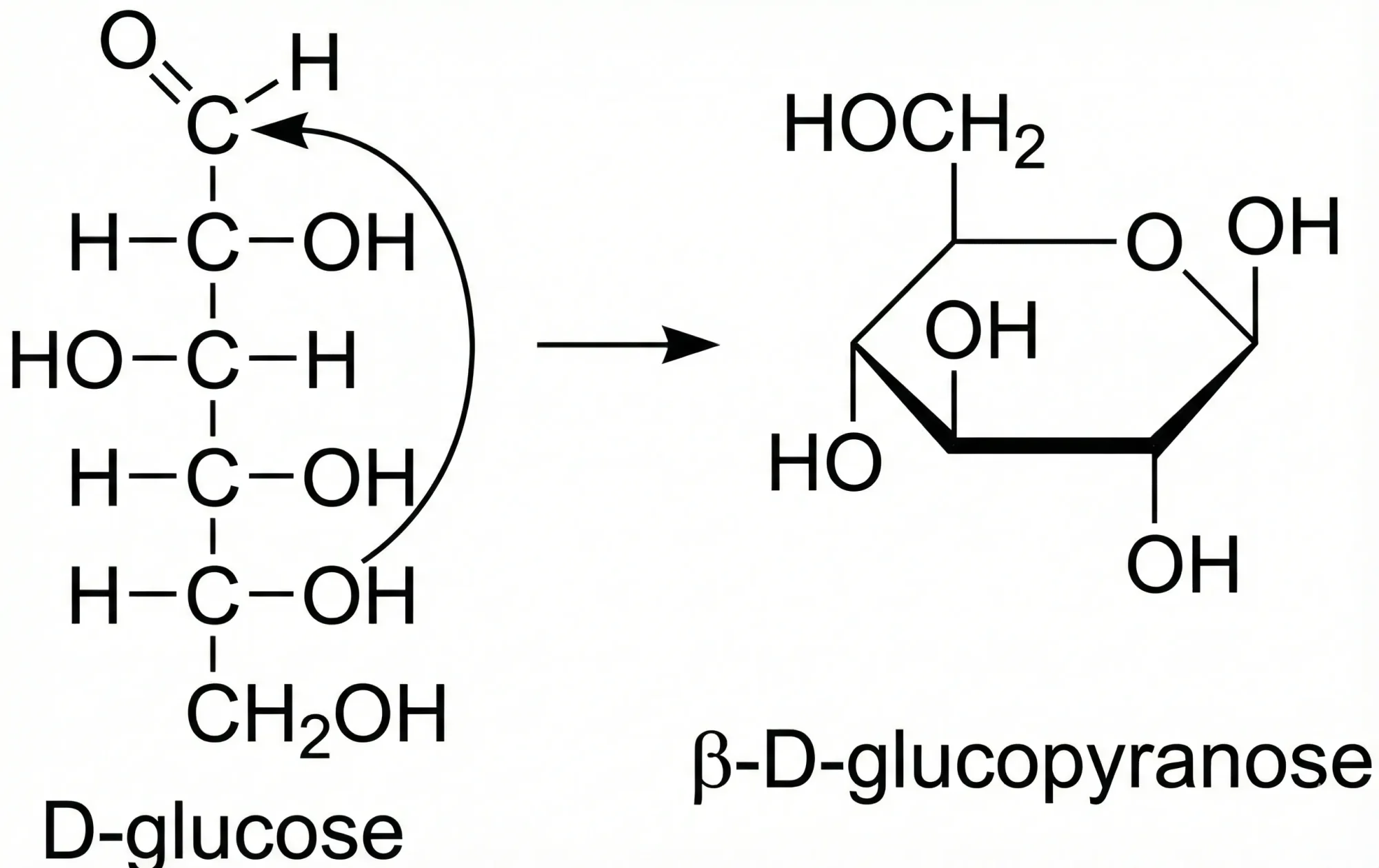
ChemReader: Automated Structure Extraction
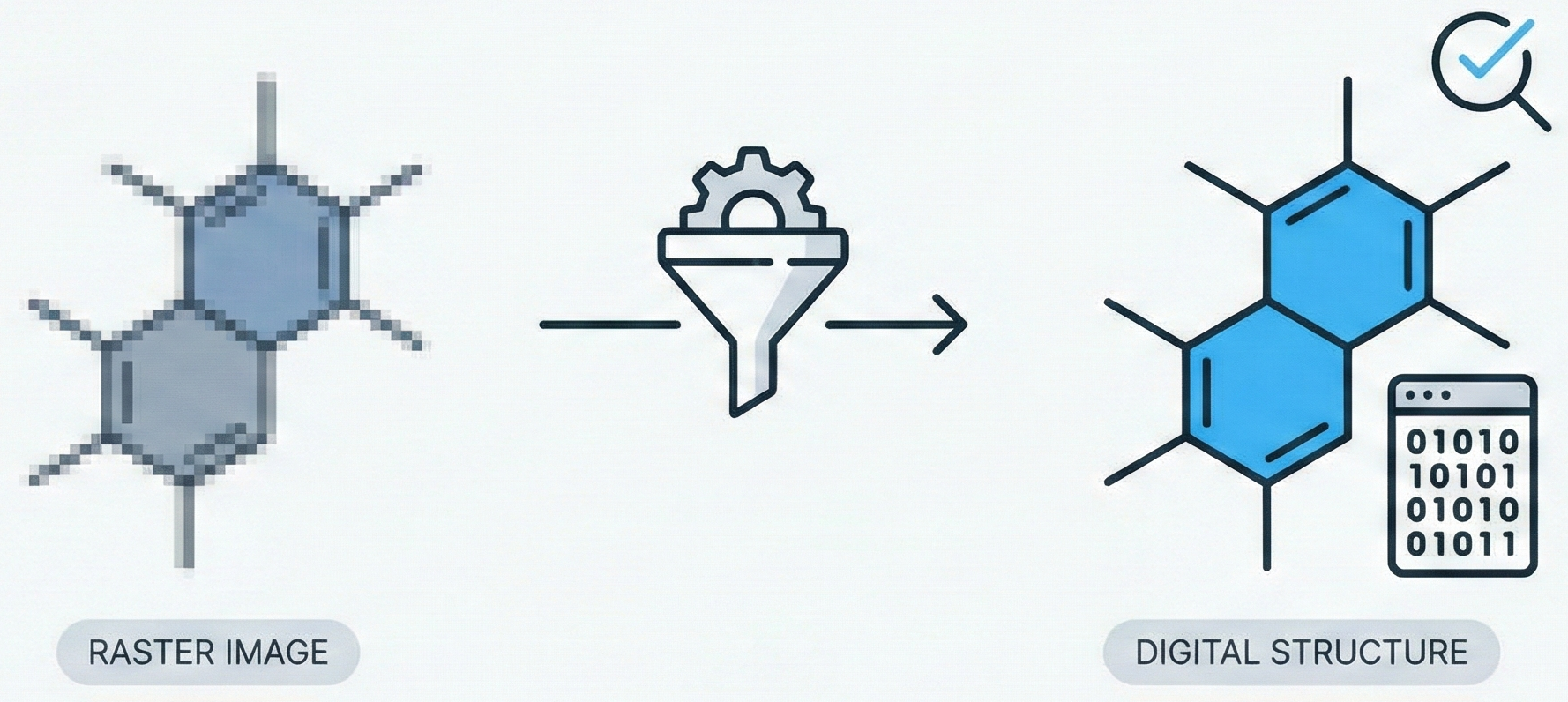
This paper presents ChemReader, a fully automated optical structure recognition tool that converts raster images of chemical diagrams into machine-readable formats. It introduces a modified Hough transform for bond detection and a chemical spell checker that improves OCR accuracy from 66% to 87%.