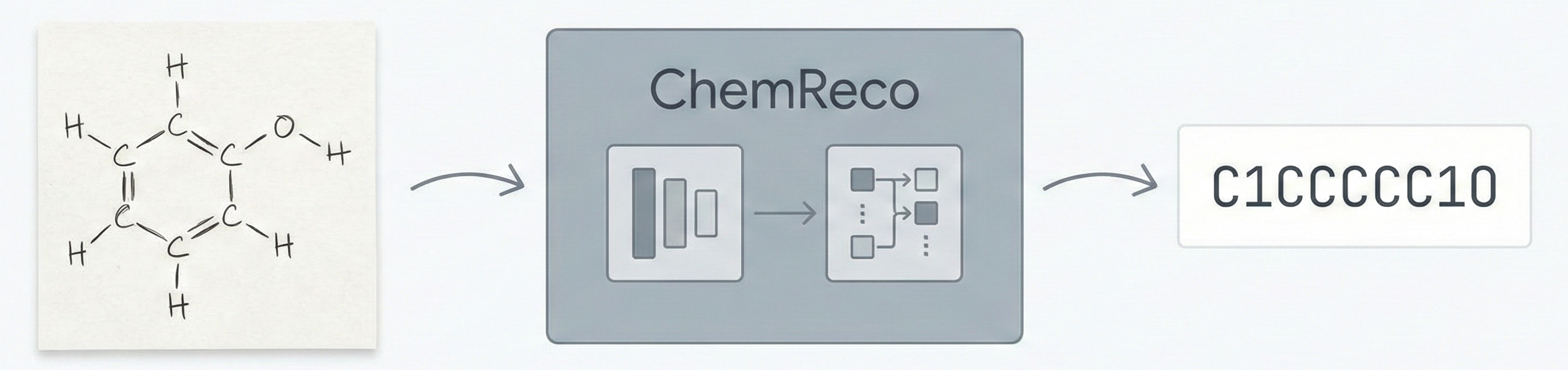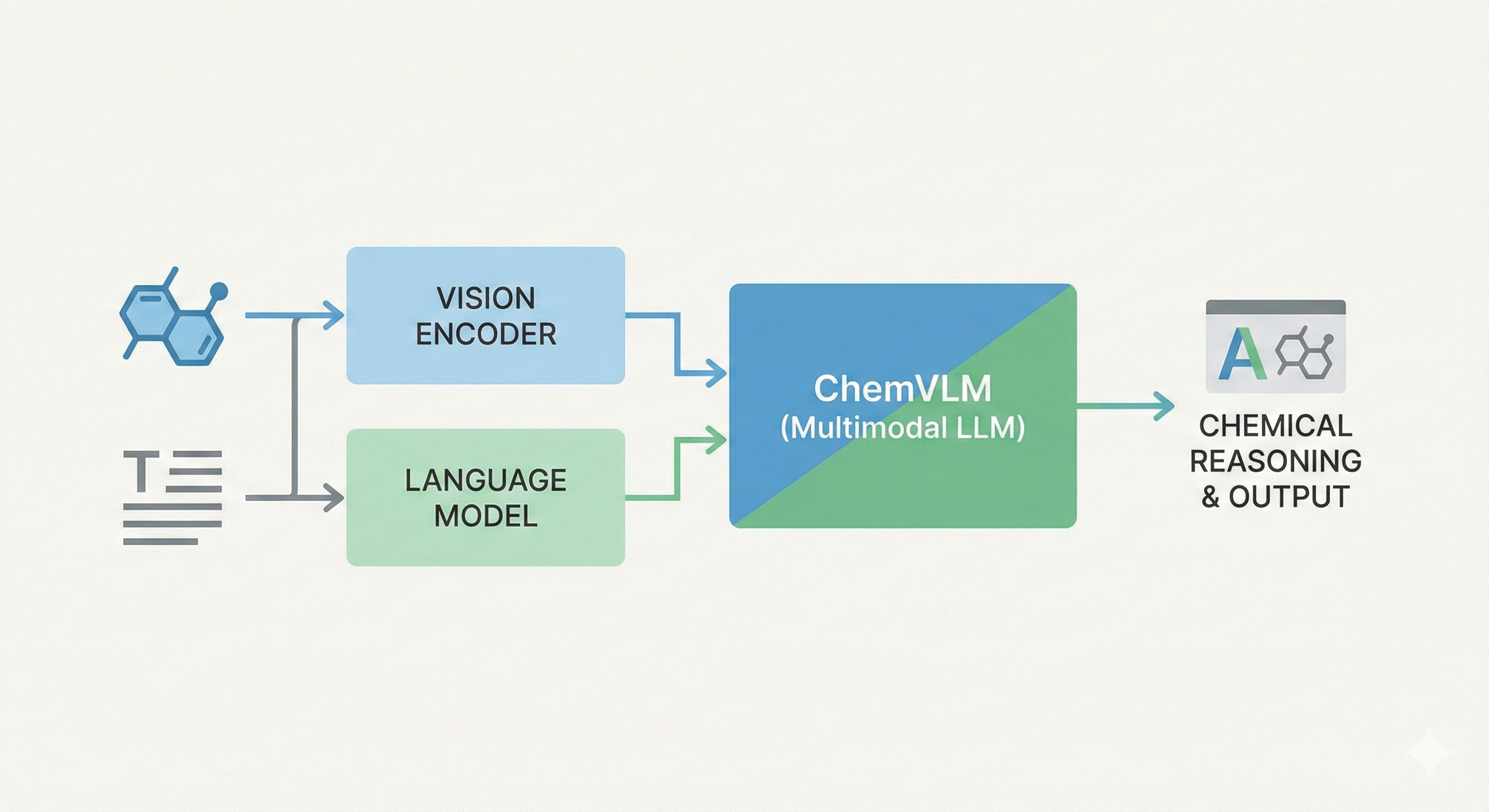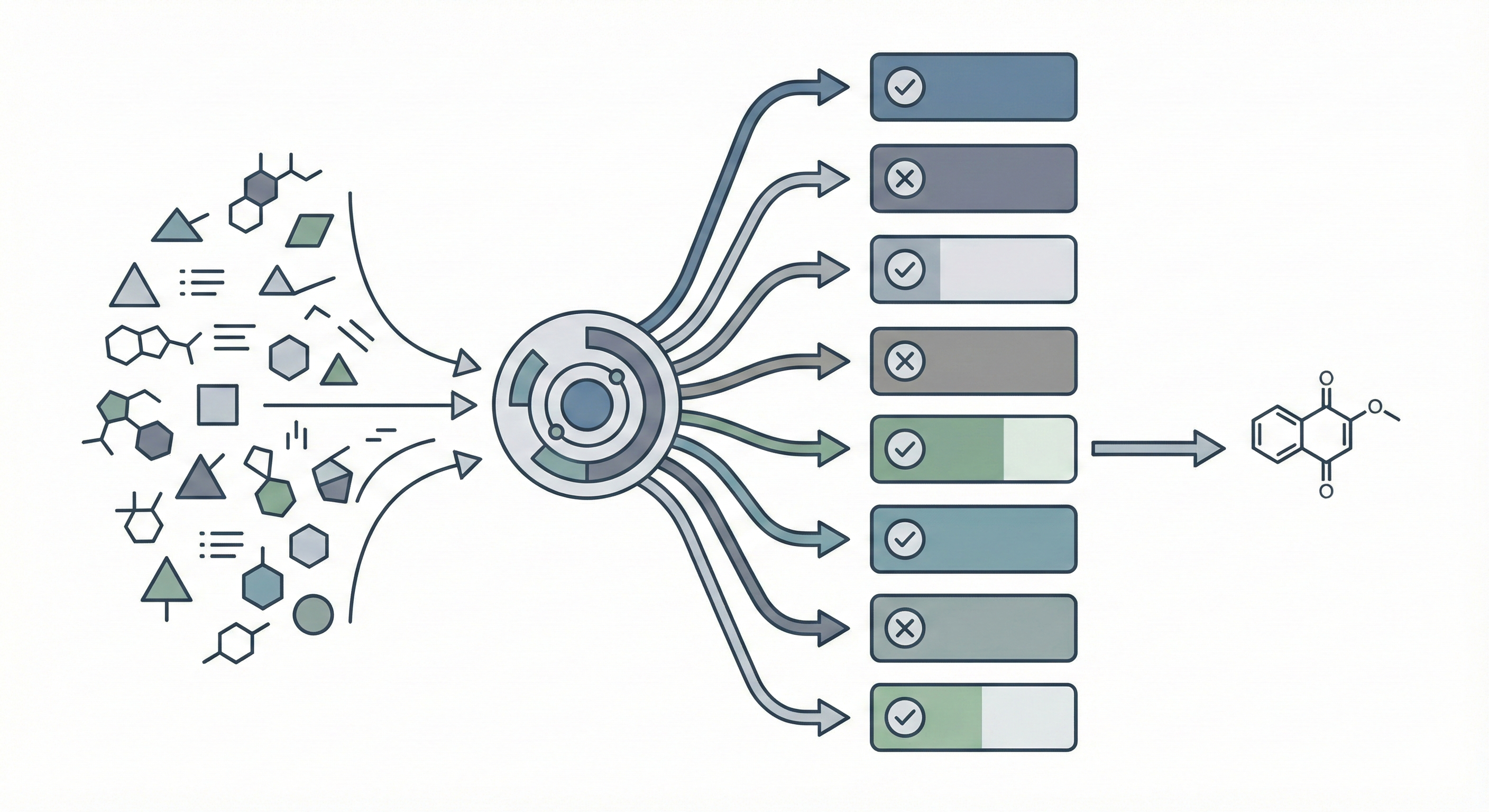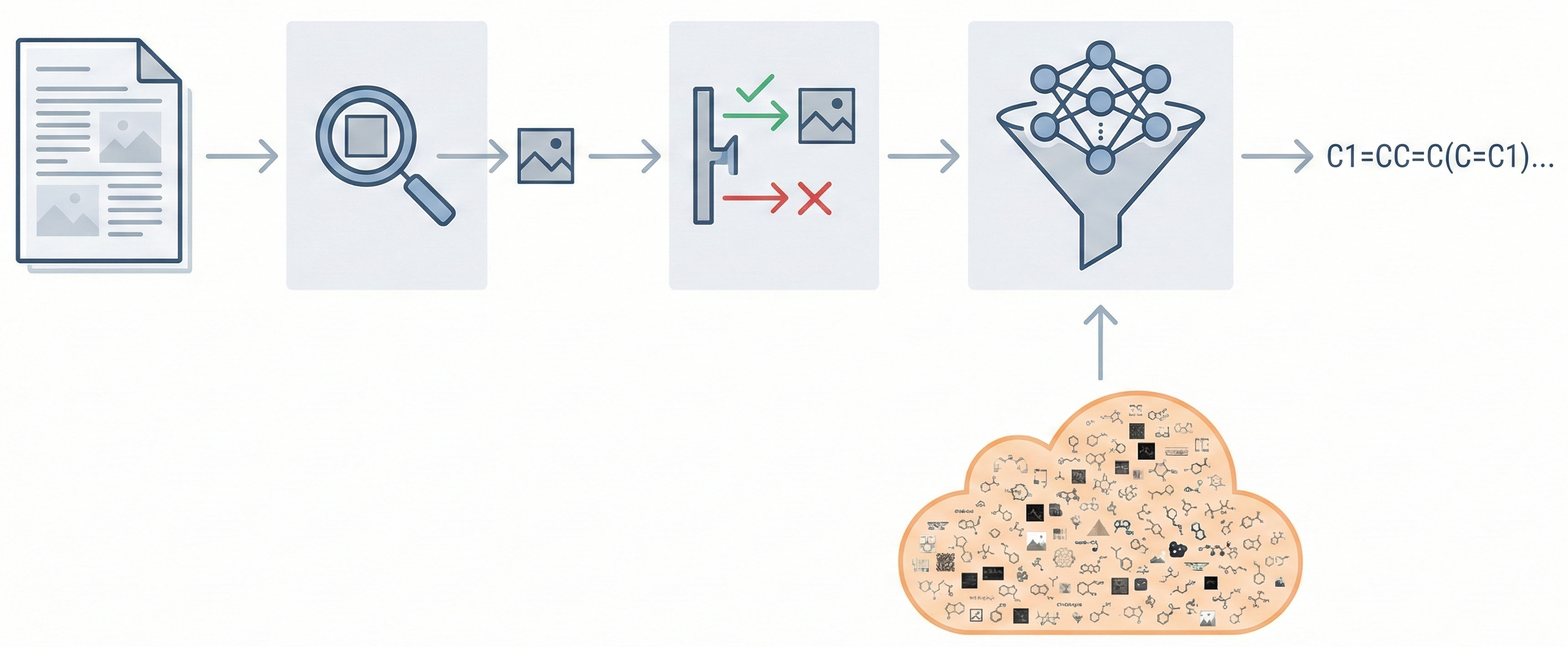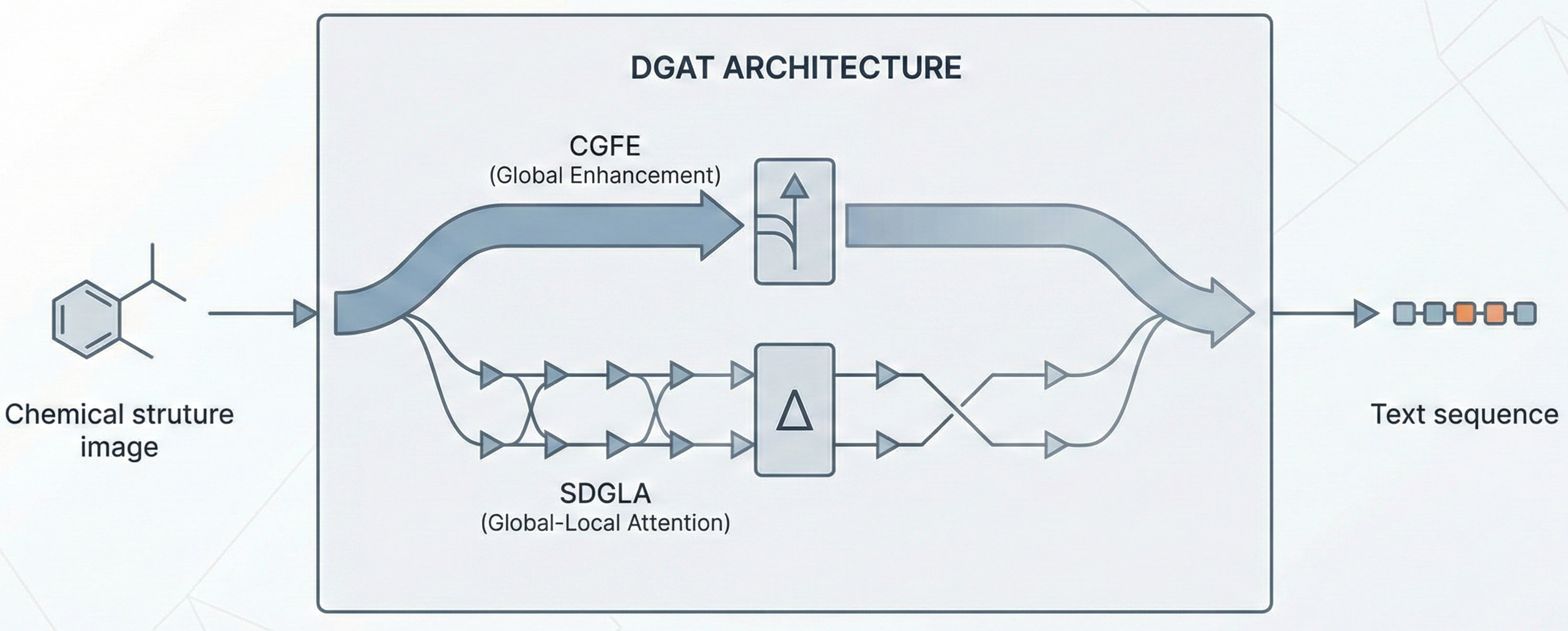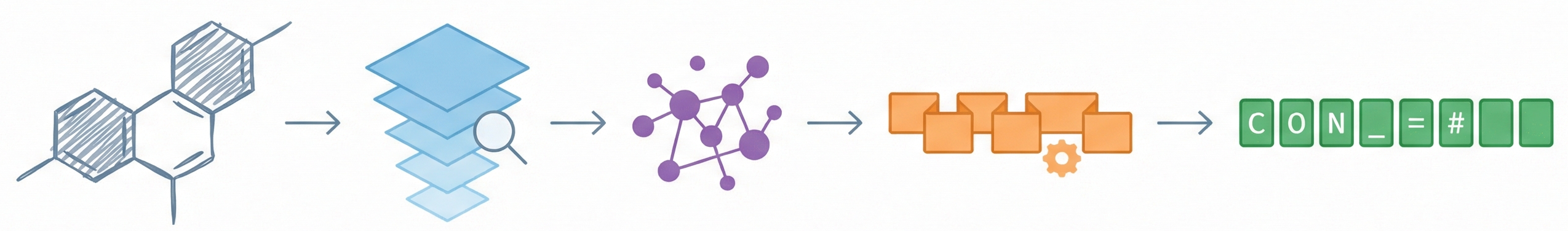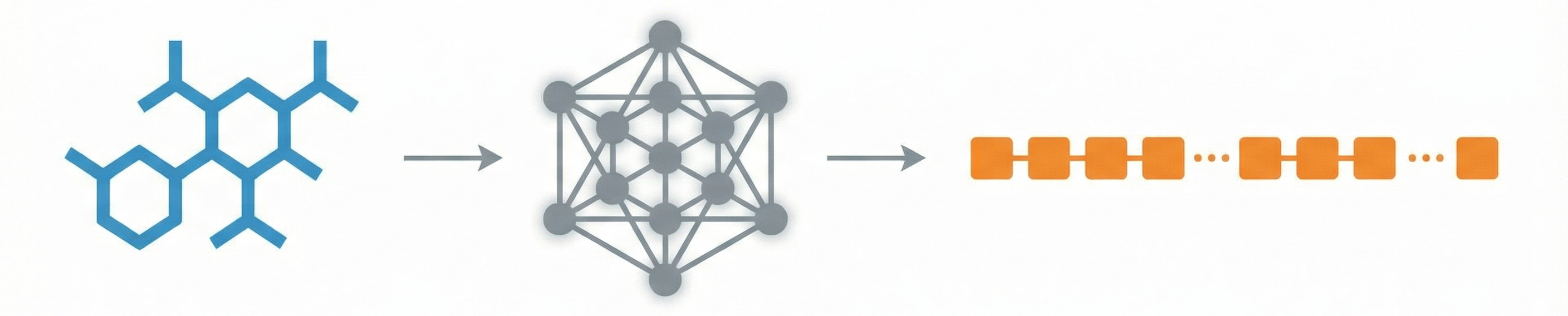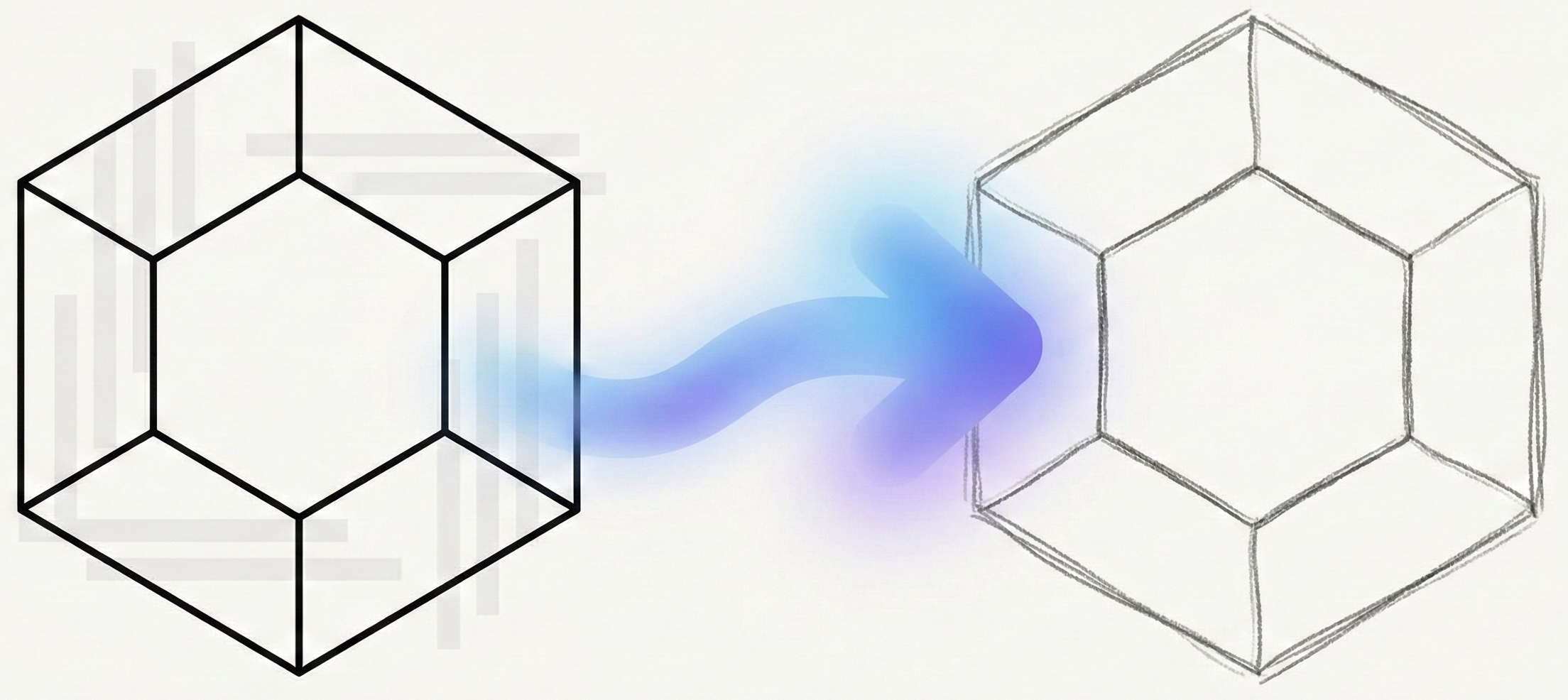
OCSAug: Diffusion-Based Augmentation for Hand-Drawn OCSR
OCSAug leverages Denoising Diffusion Probabilistic Models (DDPM) and the RePaint algorithm with custom masking to generate synthetic hand-drawn chemical structure images, significantly improving OCSR performance on benchmarks like DECIMER.