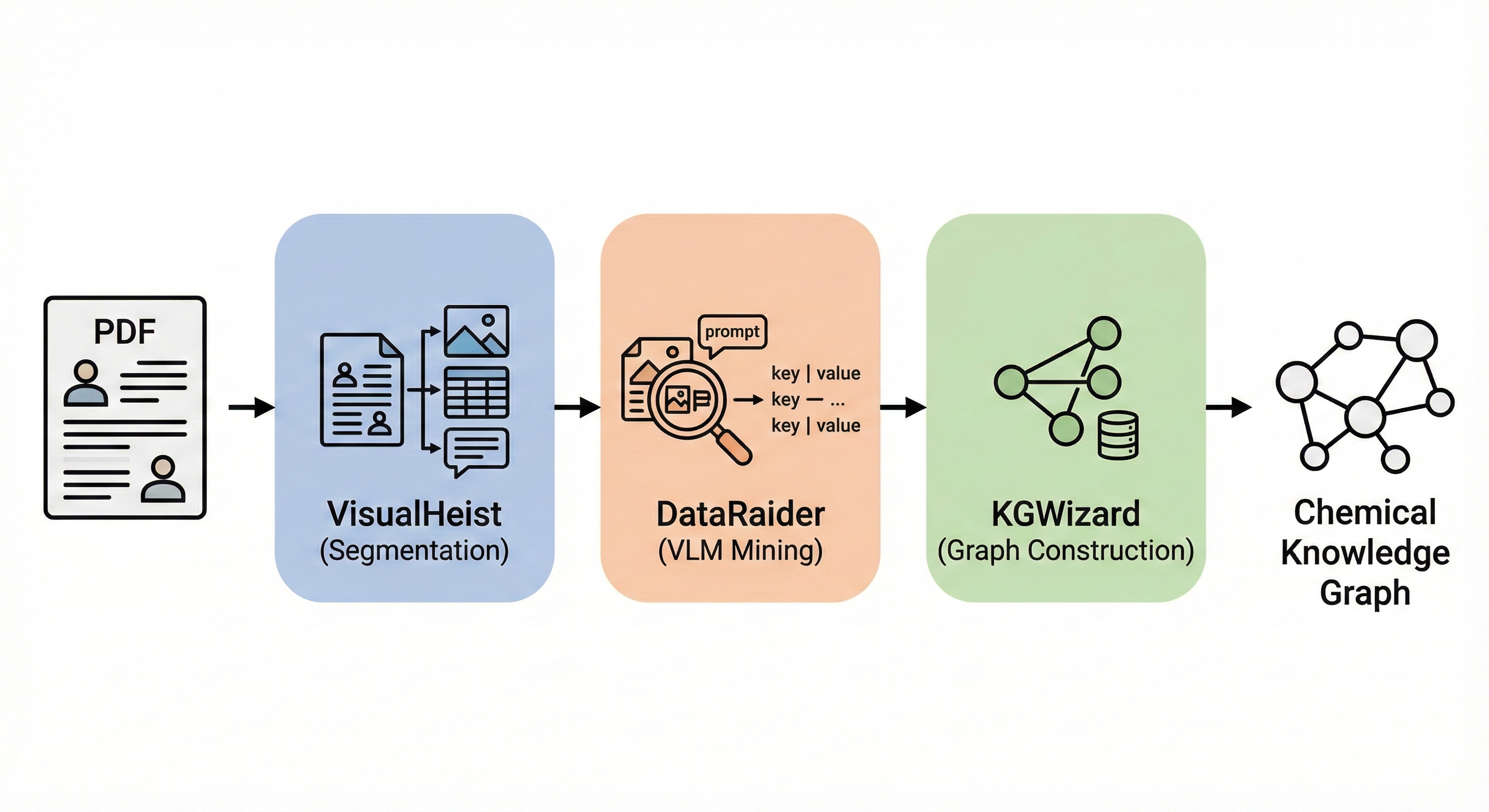
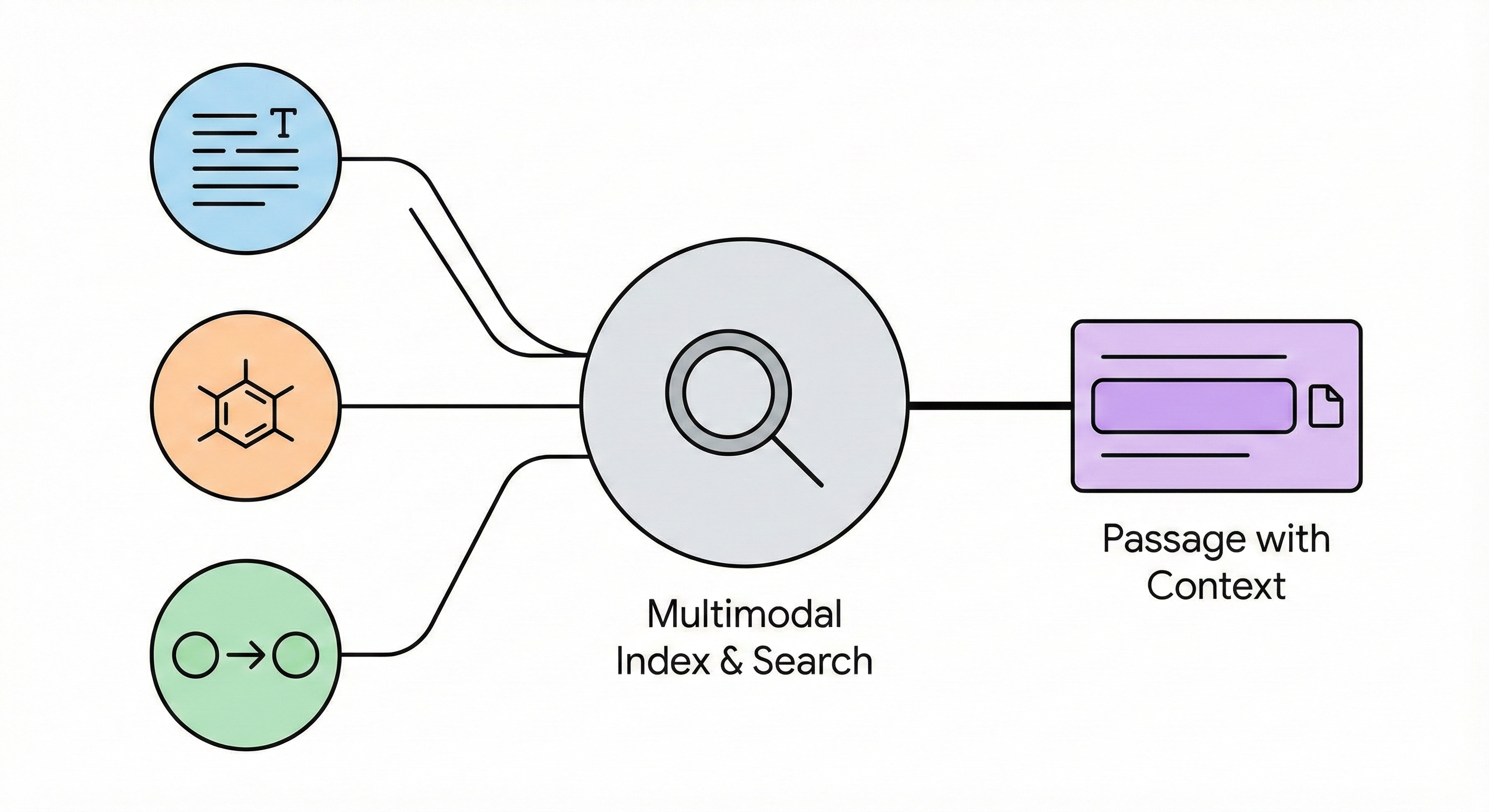
ChemBERTa-3: Open Source Training Framework
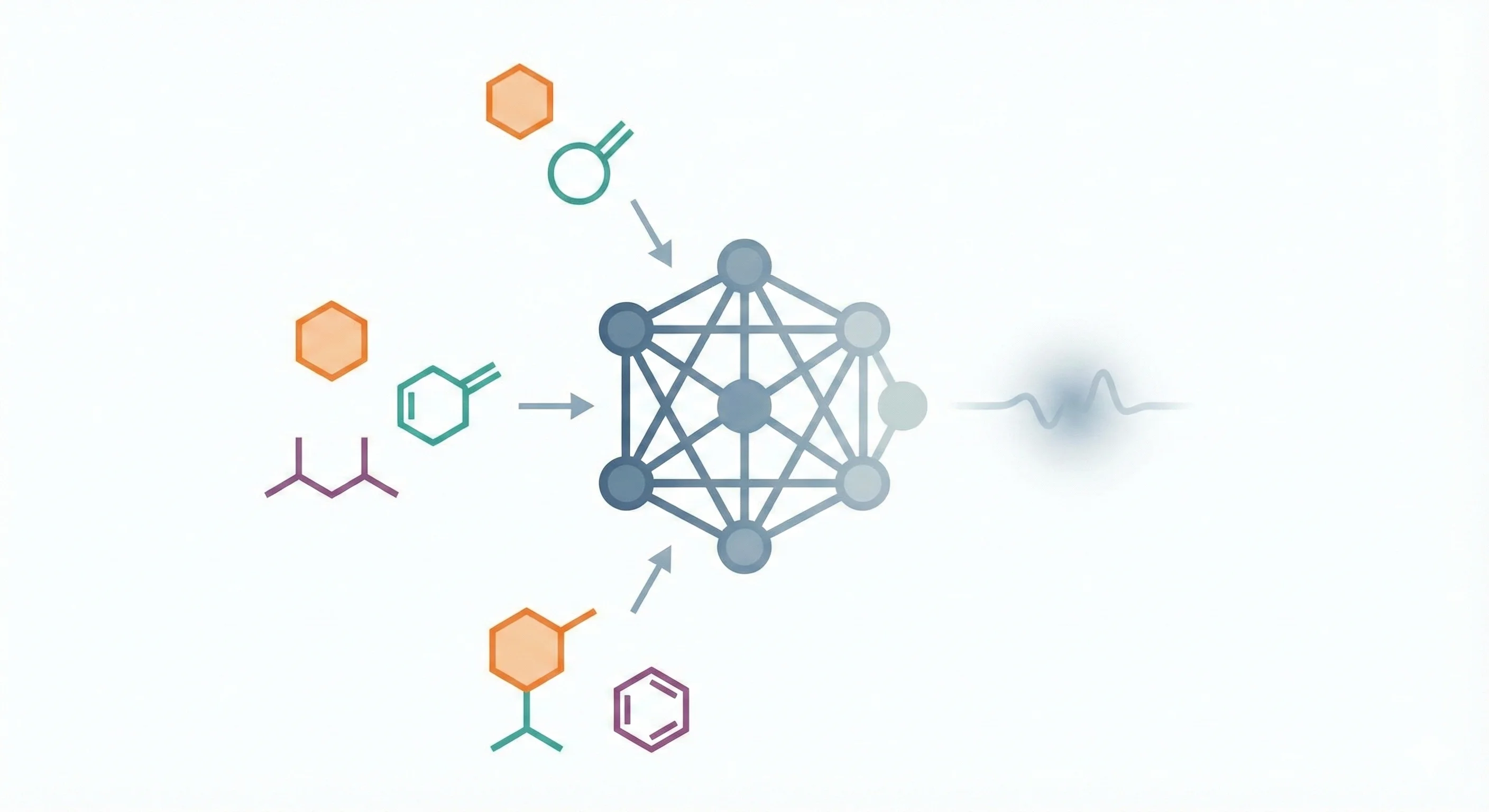
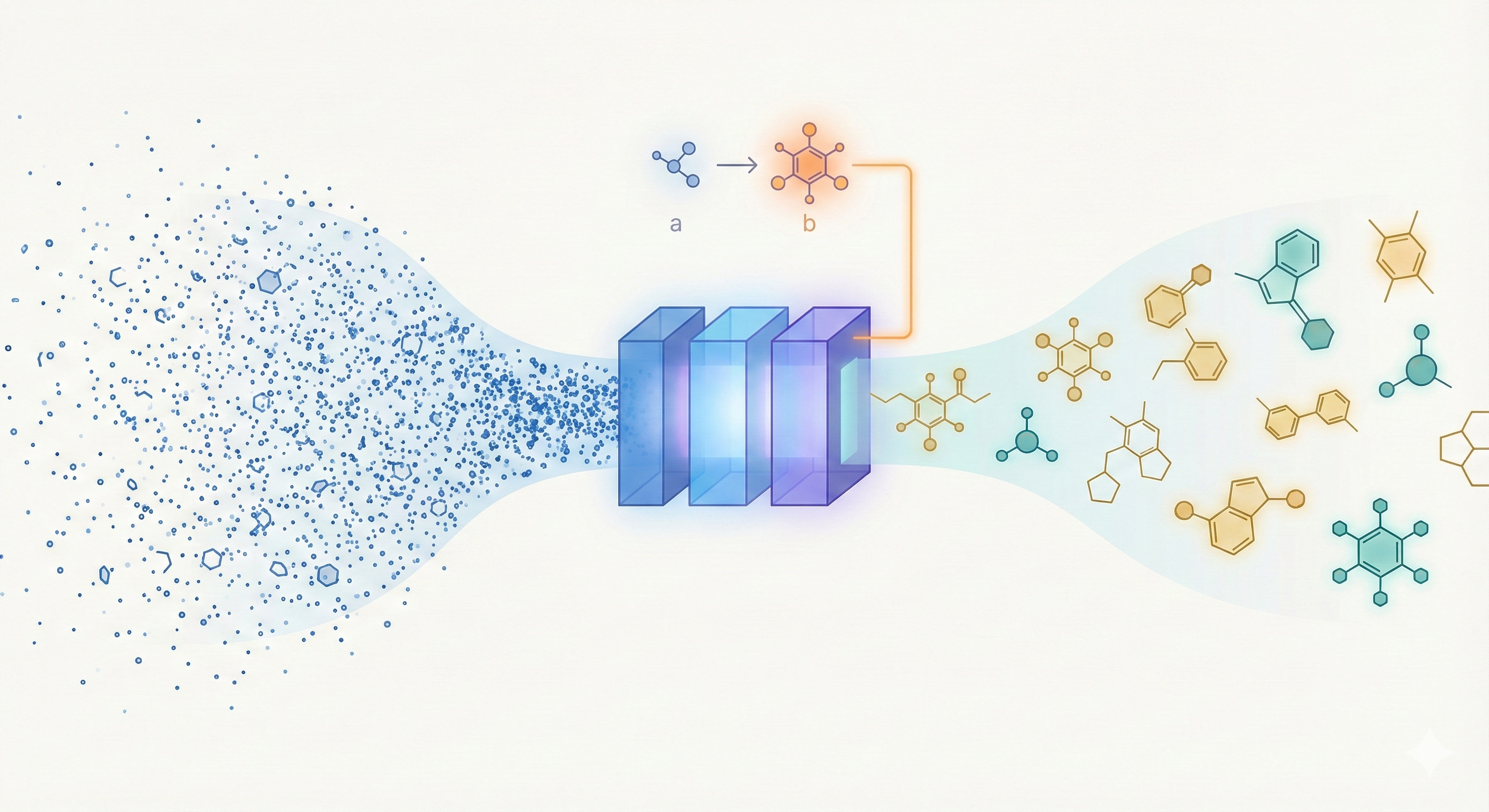
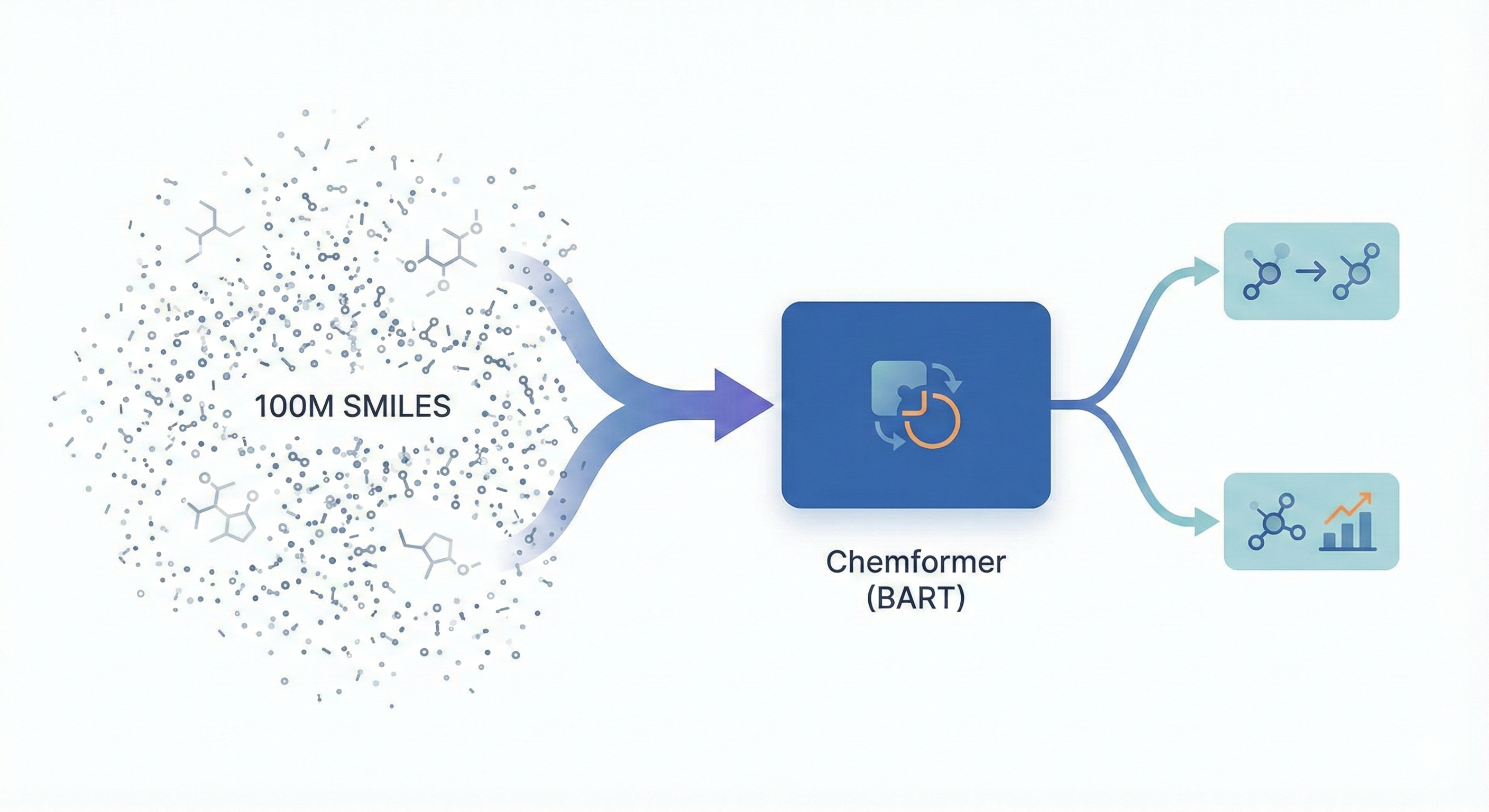
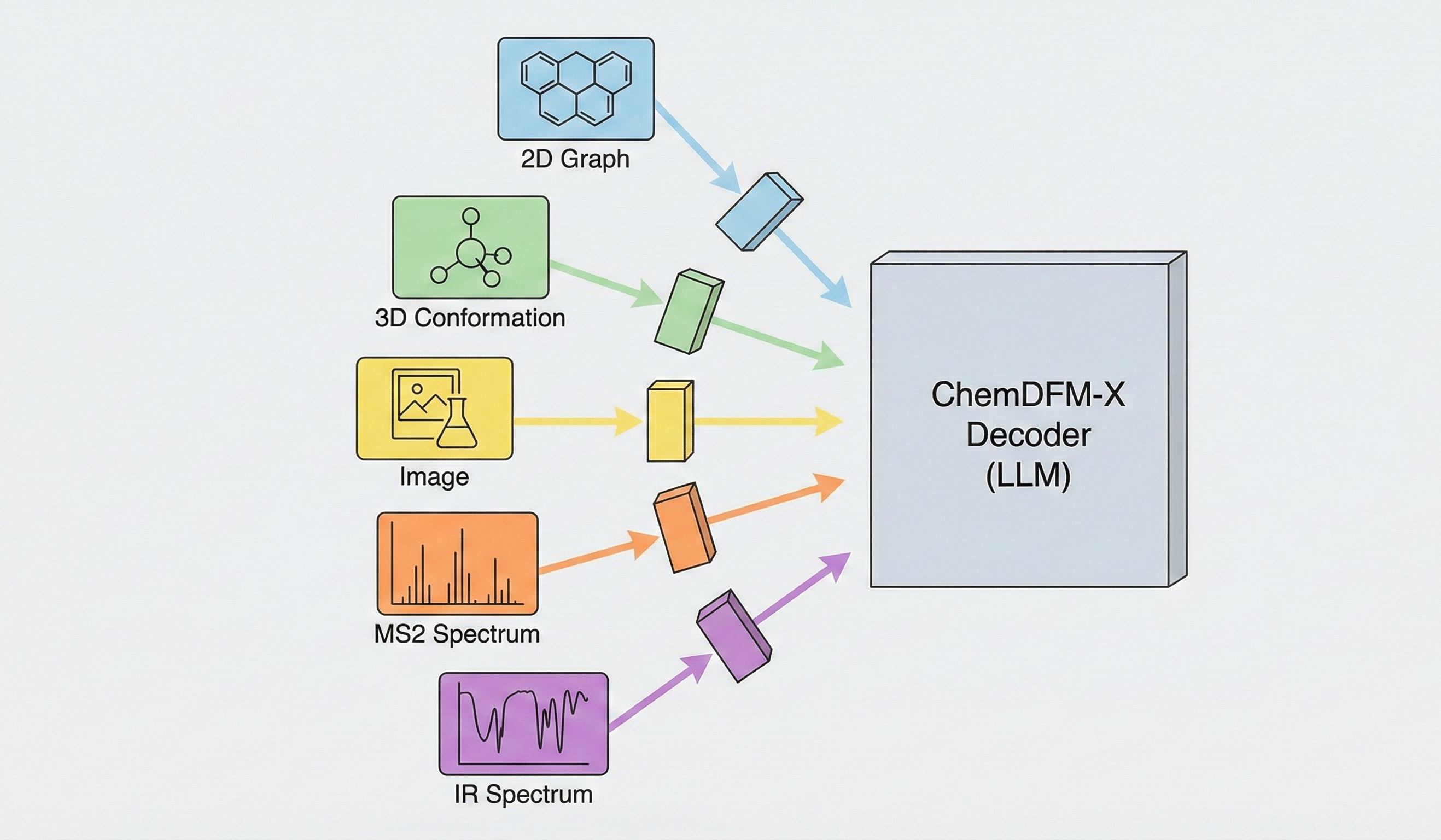
ChemBERTa-3 provides a unified, scalable infrastructure for pretraining and benchmarking chemical foundation models, addressing reproducibility gaps in previous studies like MoLFormer through standardized scaffold splitting and open-source tooling.