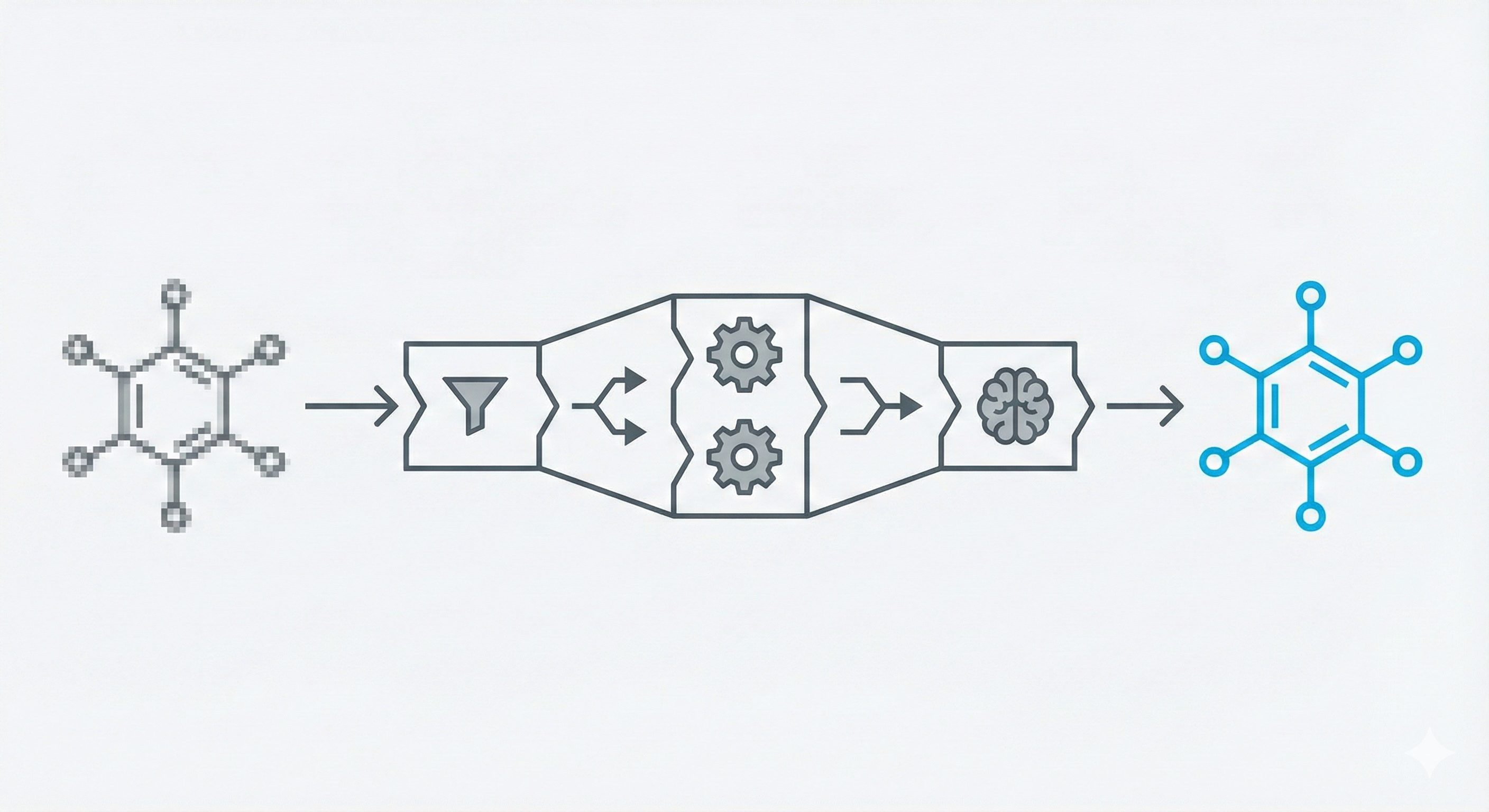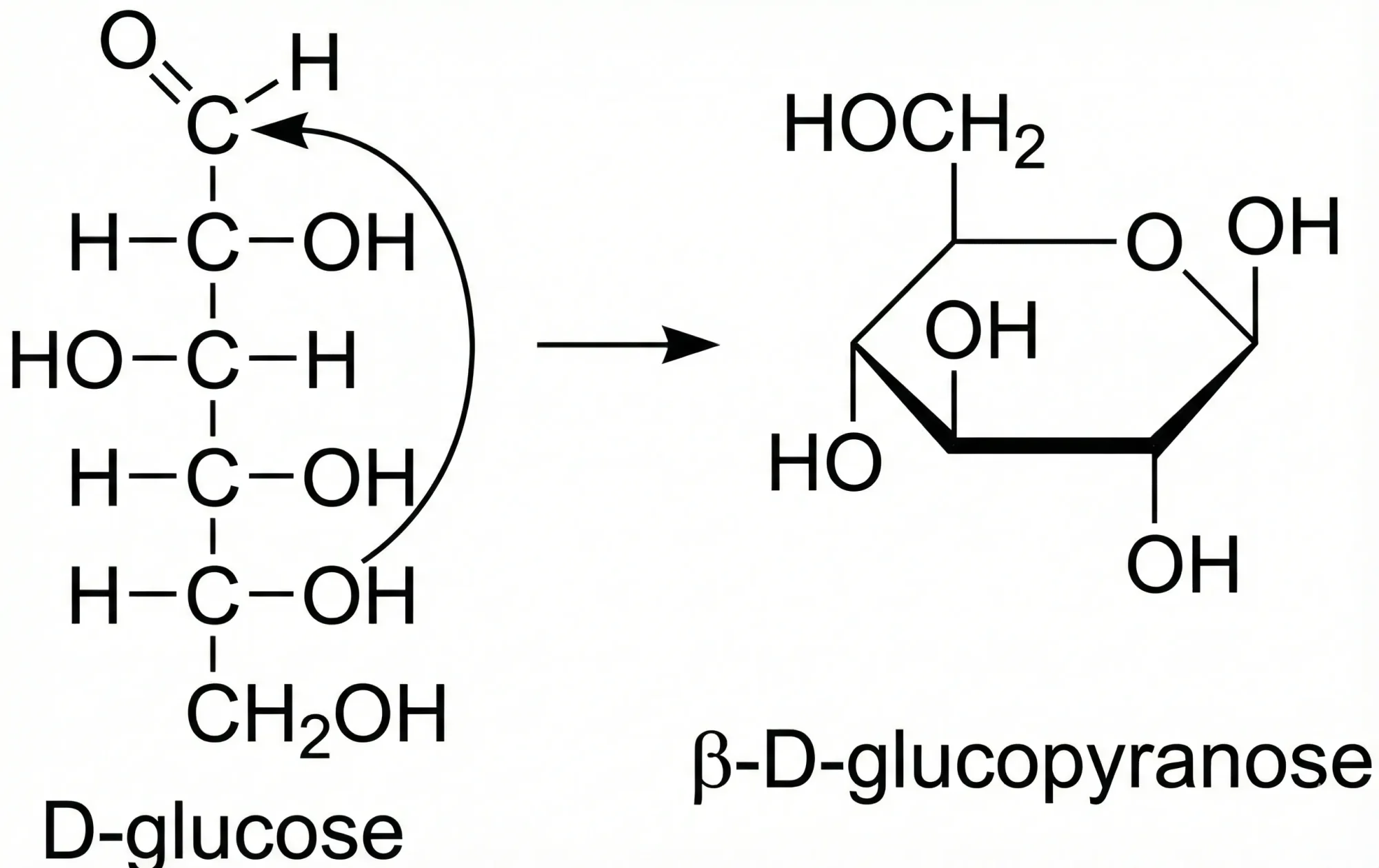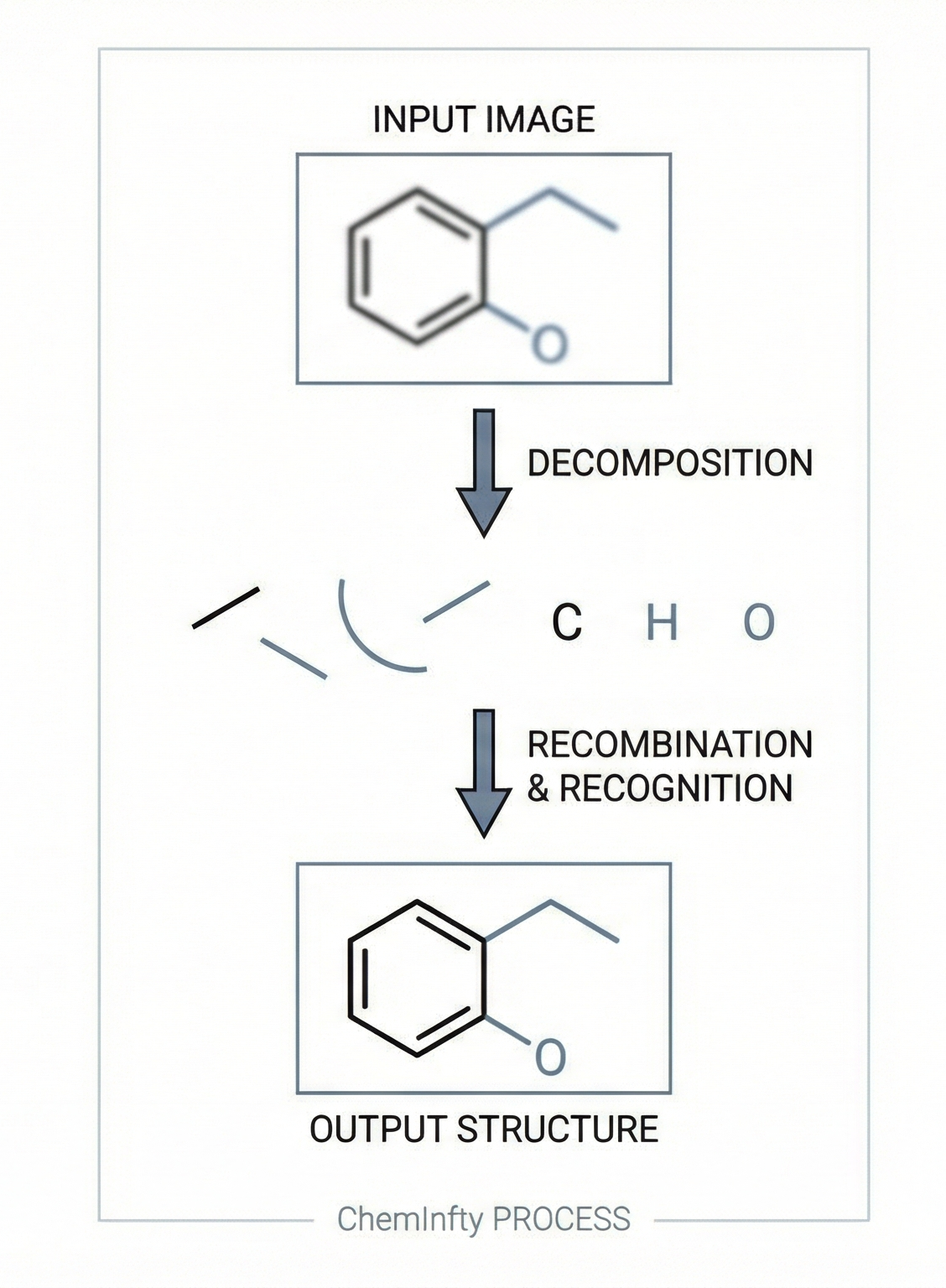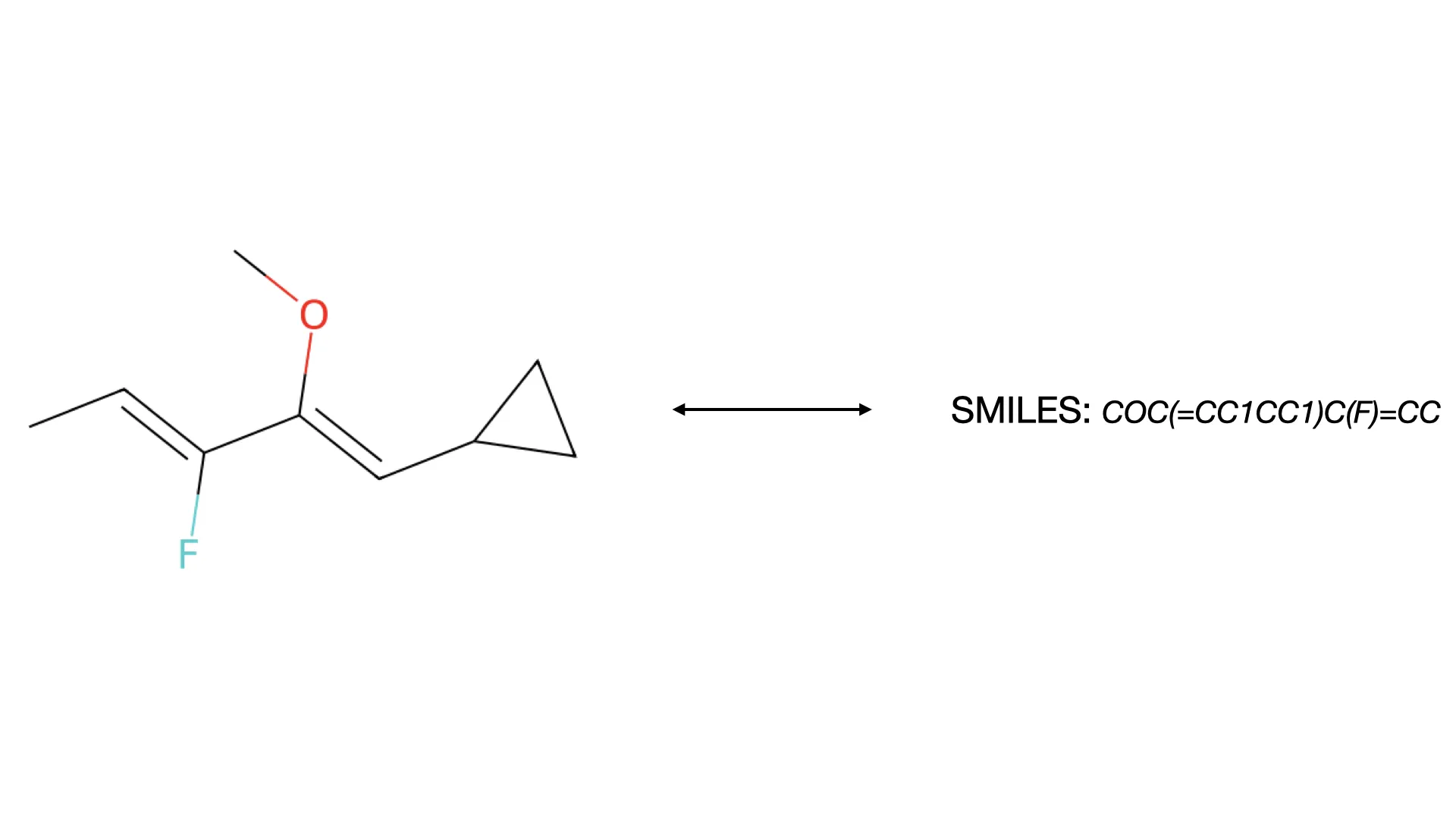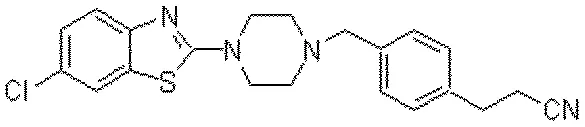
OSRA: Open Source Optical Structure Recognition
This paper presents OSRA, the first open-source utility for converting graphical chemical structures from documents into machine-readable formats (SMILES/SD). It outlines a pipeline combining existing image processing tools (ImageMagick, Potrace, GOCR) with custom heuristics for bond and atom detection, establishing a foundation for accessible chemical information extraction.