Abstract
PubMed-OCR is an OCR-centric corpus of scientific articles derived from PubMed Central Open Access PDFs. Each page image is annotated with Google Cloud Vision and released in a compact JSON schema with word-, line-, and paragraph-level bounding boxes. The corpus spans 209.5K articles (1.5M pages; ~1.3B words) and supports layout-aware modeling, coordinate-grounded QA, and evaluation of OCR-dependent pipelines. We analyze corpus characteristics (e.g., journal coverage and detected layout features) and discuss limitations, including reliance on a single OCR engine and heuristic line reconstruction. We release the data and schema to facilitate downstream research and invite extensions.
Key Contributions
- OCR-First Supervision: Unlike prior datasets for PubMed that align XML to PDFs, PubMed-OCR provides native OCR annotations (Google Cloud Vision), bypassing alignment errors and covering non-digital scanned pages.
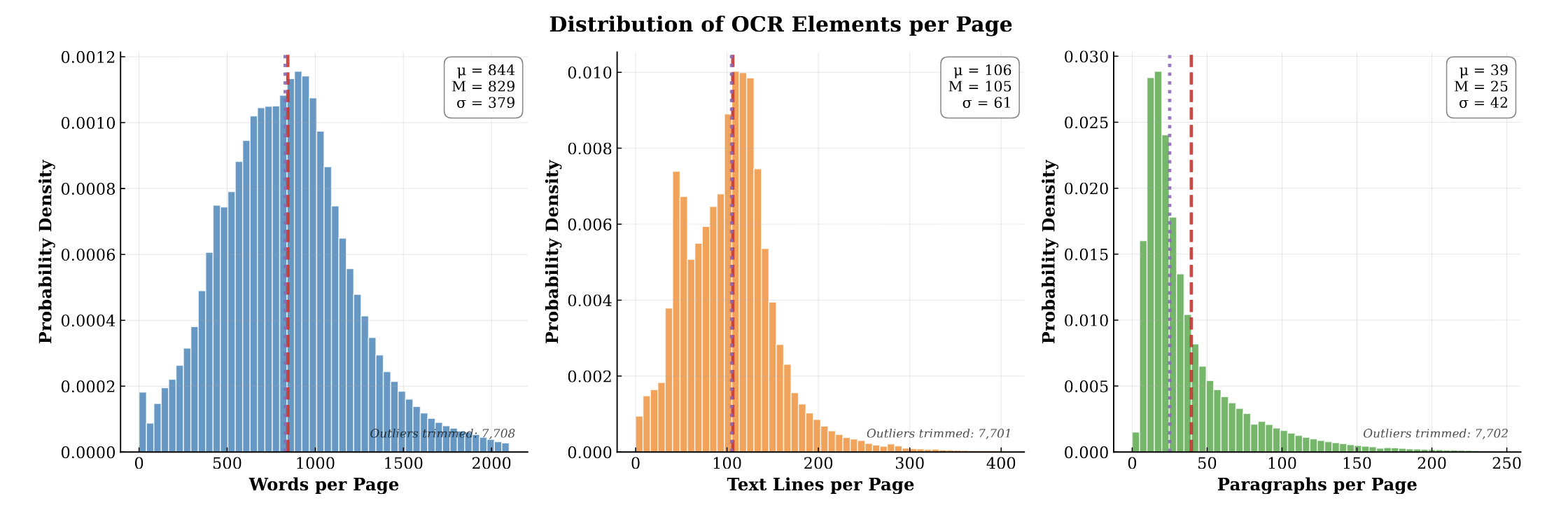
- High-Density Annotation: Offers ~1.3B words across 1.5M pages. This is significantly higher word density (10x) and line granularity (4x) than comparable industrial OCR corpora like OCR-IDL.
- Multi-Level Bounding Boxes: Includes explicit word-, line-, and paragraph-level bounding boxes to support hierarchical document understanding and layout-aware modeling. We also hope that this leads to VQA datasets with grounded answers in document layout.
- Open Access & Reproducibility: Derived strictly from the redistributable PMCOA subset, releasing both the JSON annotations and original PDFs to ensure verifiable and reproducible research.
Why This Matters
The lack of large-scale, high-quality OCR datasets with explicit geometric grounding has been a major bottleneck for training layout-aware models. By releasing PubMed-OCR, we provide the community with the dense, multi-level bounding box annotations necessary to build the next generation of document understanding systems. This dataset directly supports the development of models like GutenOCR, enabling them to learn precise token-to-pixel alignment and robust layout reasoning.
Citation
@misc{heidenreich2026pubmedocrpmcopenaccess,
title={PubMed-OCR: PMC Open Access OCR Annotations},
author={Hunter Heidenreich and Yosheb Getachew and Olivia Dinica and Ben Elliott},
year={2026},
eprint={2601.11425},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2601.11425},
}