Lately, I’ve spent a lot of time staring at datasets full of 1D molecular strings.
With time, I find I get better at recognizing functional groups and substructures like C(=O)O (carboxylic acid) or c1ccccc1 (benzene ring) in SMILES.
However, anything really complex is beyond my personal visualization capabilities.
I ran into this recently while debugging a generative model. Sometimes the grammar of the string provides the clue as to what is going wrong. Other times, actually seeing the molecule is what helps. I had a terminal full of generated strings and needed to verify their structures visually. I needed a streamlined way to generate these images locally. A lightweight script turns that text into a properly formatted image directly from the terminal.
SMILES vs. SELFIES
There are two primary string representations you will encounter in modern cheminformatics:
- SMILES: The industry standard. It uses simple rules (
Cfor carbon,=for double bonds, parentheses for branches). It is compact and machine-parseable. However, random SMILES strings are often invalid (e.g., unclosed rings or invalid valences). - SELFIES: Designed specifically for machine learning. It is a robust representation where every string corresponds to a valid molecular graph. This makes it ideal for generative models. Note that it is more verbose than SMILES.
I often need to visualize both formats. Let’s build a single, robust Python tool to handle them.
The Quick Win: Native RDKit
If you just need a quick image from a SMILES string and don’t care about the image dimensions or adding a legend, RDKit can do this in three lines:
from rdkit import Chem
from rdkit.Chem import Draw
mol = Chem.MolFromSmiles("CCO")
Draw.MolToFile(mol, "ethanol.png")
The native RDKit method is fast for quick checks. However, custom rendering provides necessary control over image dimensions, formula subscripts, and handling multiple input formats like SELFIES.
Building a Custom Renderer for Precise Control
Let’s build a more robust tool using RDKit for chemical processing, the selfies library for decoding, and PIL for high-quality image manipulation.
Core Dependencies
import selfies as sf
from rdkit import Chem
from rdkit.Chem import Draw, rdDepictor, rdMolDescriptors
from PIL import Image, ImageDraw, ImageFont
RDKit handles the chemical logic, selfies translates SELFIES to SMILES, and PIL gives us fine control over the final image appearance.
The Main Conversion Function
Here is the core conversion logic. Notice the addition of Python type hints to ensure our code is robust and maintainable.
def string_to_png(mol_string: str, output_file: str, size: int = 500, is_selfies: bool = False) -> None:
"""Generates a 2D molecule image with a chemical formula legend from SMILES or SELFIES."""
# Decode SELFIES to SMILES if necessary
if is_selfies:
try:
smiles = sf.decoder(mol_string)
except Exception as e:
raise ValueError(f"Invalid SELFIES string: {mol_string}") from e
else:
smiles = mol_string
mol = Chem.MolFromSmiles(smiles)
if not mol:
raise ValueError(f"Could not generate molecule from SMILES: {smiles}")
# Generate 2D coordinates and formula
rdDepictor.Compute2DCoords(mol)
formula = rdMolDescriptors.CalcMolFormula(mol)
# Render the molecule
img = Draw.MolToImage(mol, size=(size, size)).convert("RGBA")
# Create a canvas with extra space at the bottom for the legend
legend_height = int(size * 0.1)
canvas = Image.new("RGBA", (size, size + legend_height), "white")
canvas.paste(img, (0, 0))
draw = ImageDraw.Draw(canvas)
# Define dynamic font sizes
font_reg = get_font(int(size * 0.03))
font_sub = get_font(int(size * 0.02))
# Draw the legend
x = int(size * 0.02)
y = size + int(size * 0.02)
# Draw "Formula: " label
draw.text((x, y), "Formula: ", fill="black", font=font_reg)
x += draw.textlength("Formula: ", font=font_reg)
# Draw formula with subscript handling for numbers
for char in formula:
# Use smaller font and lower y-offset for numbers (subscripts)
font = font_sub if char.isdigit() else font_reg
y_offset = int(size * 0.005) if char.isdigit() else 0
draw.text((x, y + y_offset), char, fill="black", font=font)
x += draw.textlength(char, font=font)
# Draw original string
label = "SELFIES" if is_selfies else "SMILES"
draw.text((x, y), f" | {label}: {mol_string}", fill="black", font=font_reg)
canvas.save(output_file)
print(f"Saved: {output_file}")
This function handles everything: SELFIES decoding, validation, coordinate generation, image creation, and legend drawing.
Font Handling
We need a helper to handle fonts robustly across systems:
def get_font(size: int, font_name: str = "arial.ttf"):
"""Attempts to load a TTF font, falls back to default if unavailable."""
try:
return ImageFont.truetype(font_name, size)
except IOError:
return ImageFont.load_default()
Examples in Action
Let’s see the tool in action with some common molecules, comparing the SMILES and SELFIES inputs.
Simple Molecules
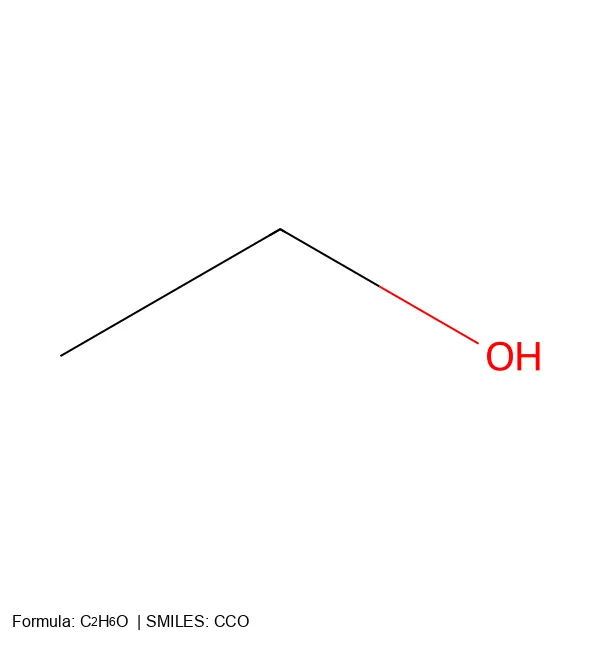
CCO, while the SELFIES is [C][C][O].Aromatic Compounds
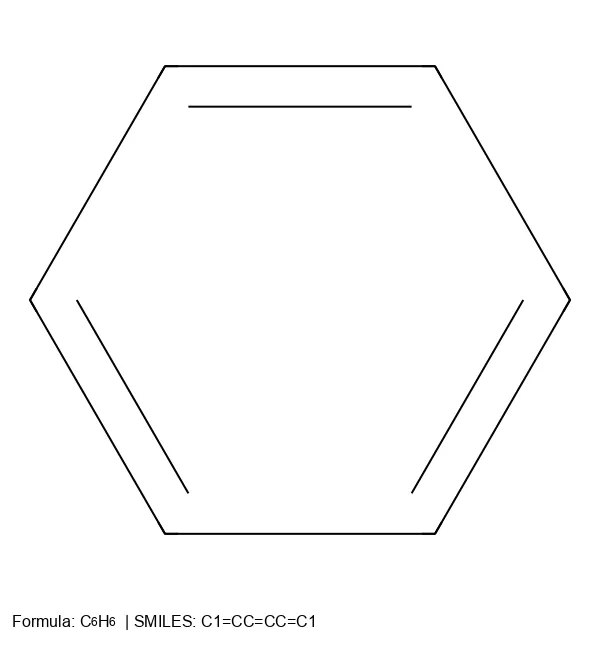
C1=CC=CC=C1), while SELFIES uses explicit tokens ([C][=C][C][=C][C][=C][Ring1][=Branch1]).Complex Pharmaceuticals
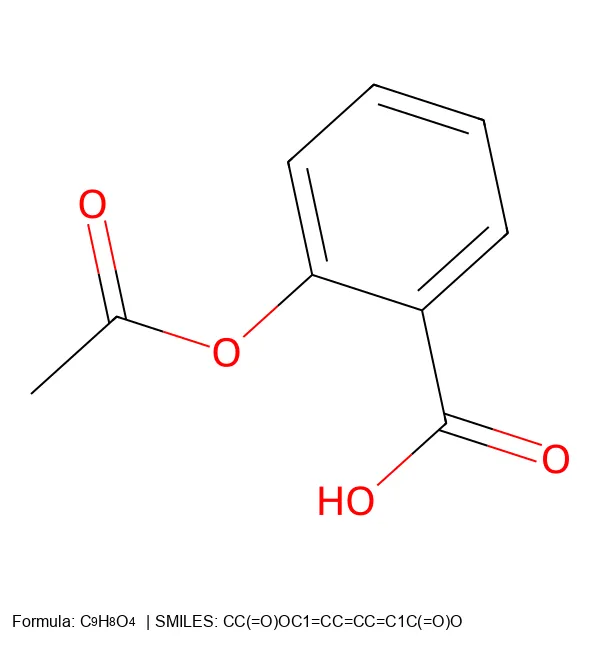
Going Further: Vector Graphics (SVG)
Use vector graphics (SVG/PDF) for true publication-quality figures. Vector graphics scale infinitely without pixelation.
RDKit handles this natively with rdMolDraw2D:
from rdkit import Chem
from rdkit.Chem.Draw import rdMolDraw2D
def string_to_svg(mol_string: str, output_file: str, size: int = 500, is_selfies: bool = False) -> None:
"""Generates a 2D molecule SVG image."""
if is_selfies:
try:
mol_string = sf.decoder(mol_string)
except Exception as e:
raise ValueError(f"Invalid SELFIES string: {mol_string}") from e
mol = Chem.MolFromSmiles(mol_string)
if not mol:
raise ValueError(f"Invalid string: {mol_string}")
rdDepictor.Compute2DCoords(mol)
d = rdMolDraw2D.MolDraw2DSVG(size, size)
d.DrawMolecule(mol)
d.FinishDrawing()
with open(output_file, "w") as f:
f.write(d.GetDrawingText())
print(f"Saved: {output_file}")
This provides a perfect vector image. Note that this method omits the custom PIL-based legend. Choose the right tool for the job: PNG for quick checks and slides, SVG for journal submissions.
Command-Line Interface
The tool uses Python’s standard argparse library for a robust command-line interface. It automatically detects if you want an SVG based on the file extension and includes a --selfies flag.
# Basic SMILES usage
python mol2img.py "CCO" -o ethanol.png
# SELFIES usage
python mol2img.py "[C][C][O]" -o ethanol.png --selfies
# Generate SVG for publication
python mol2img.py "CCO" -o ethanol.svg
Download the Complete Script
You can copy the complete mol2img.py script directly from the code block below.
Installation and Setup
Before using the script, install the required dependencies:
pip install rdkit pillow selfies
Complete Script
Click to expand the complete mol2img.py script
import argparse
import sys
import os
import selfies as sf
from rdkit import Chem
from rdkit.Chem import Draw, rdDepictor, rdMolDescriptors
from rdkit.Chem.Draw import rdMolDraw2D
from PIL import Image, ImageDraw, ImageFont
def get_font(size: int, font_name: str = "arial.ttf"):
"""Attempts to load a TTF font, falls back to default if unavailable."""
try:
return ImageFont.truetype(font_name, size)
except IOError:
return ImageFont.load_default()
def string_to_svg(mol_string: str, output_file: str, size: int = 500, is_selfies: bool = False) -> None:
"""Generates a 2D molecule SVG image."""
if is_selfies:
try:
mol_string = sf.decoder(mol_string)
except Exception as e:
raise ValueError(f"Invalid SELFIES string: {mol_string}") from e
mol = Chem.MolFromSmiles(mol_string)
if not mol:
raise ValueError(f"Invalid string: {mol_string}")
rdDepictor.Compute2DCoords(mol)
d = rdMolDraw2D.MolDraw2DSVG(size, size)
d.DrawMolecule(mol)
d.FinishDrawing()
with open(output_file, "w") as f:
f.write(d.GetDrawingText())
print(f"Saved: {output_file}")
def string_to_png(mol_string: str, output_file: str, size: int = 500, is_selfies: bool = False) -> None:
"""Generates a 2D molecule image with a chemical formula legend."""
if is_selfies:
try:
smiles = sf.decoder(mol_string)
except Exception as e:
raise ValueError(f"Invalid SELFIES string: {mol_string}") from e
else:
smiles = mol_string
mol = Chem.MolFromSmiles(smiles)
if not mol:
raise ValueError(f"Could not generate molecule from string: {mol_string}")
# Generate 2D coordinates and formula
rdDepictor.Compute2DCoords(mol)
formula = rdMolDescriptors.CalcMolFormula(mol)
# Render the molecule
img = Draw.MolToImage(mol, size=(size, size)).convert("RGBA")
# Create a canvas with extra space at the bottom for the legend
legend_height = int(size * 0.1)
canvas = Image.new("RGBA", (size, size + legend_height), "white")
canvas.paste(img, (0, 0))
draw = ImageDraw.Draw(canvas)
# Define dynamic font sizes
font_reg = get_font(int(size * 0.03))
font_sub = get_font(int(size * 0.02))
# Draw the legend
x = int(size * 0.02)
y = size + int(size * 0.02)
# Draw "Formula: " label
draw.text((x, y), "Formula: ", fill="black", font=font_reg)
x += draw.textlength("Formula: ", font=font_reg)
# Draw formula with subscript handling for numbers
for char in formula:
# Use smaller font and lower y-offset for numbers (subscripts)
font = font_sub if char.isdigit() else font_reg
y_offset = int(size * 0.005) if char.isdigit() else 0
draw.text((x, y + y_offset), char, fill="black", font=font)
x += draw.textlength(char, font=font)
# Draw original string
label = "SELFIES" if is_selfies else "SMILES"
draw.text((x, y), f" | {label}: {mol_string}", fill="black", font=font_reg)
canvas.save(output_file)
print(f"Saved: {output_file}")
if __name__ == "__main__":
parser = argparse.ArgumentParser(description="Convert a SMILES or SELFIES string to a 2D molecular image.")
parser.add_argument("string", help="The molecular string to convert")
parser.add_argument("-o", "--output", default="molecule.png", help="Output filename (default: molecule.png)")
parser.add_argument("--size", type=int, default=500, help="Image width/height in pixels (default: 500)")
parser.add_argument("--svg", action="store_true", help="Force SVG output (overrides filename extension)")
parser.add_argument("--selfies", action="store_true", help="Treat the input string as SELFIES.")
args = parser.parse_args()
try:
# Determine format based on flag or file extension
is_svg = args.svg or args.output.lower().endswith(".svg")
if is_svg:
# Ensure extension is correct if not present
if not args.output.lower().endswith(".svg"):
args.output = os.path.splitext(args.output)[0] + ".svg"
string_to_svg(args.string, args.output, args.size, args.selfies)
else:
string_to_png(args.string, args.output, args.size, args.selfies)
except Exception as e:
print(f"Error: {e}")
sys.exit(1)
