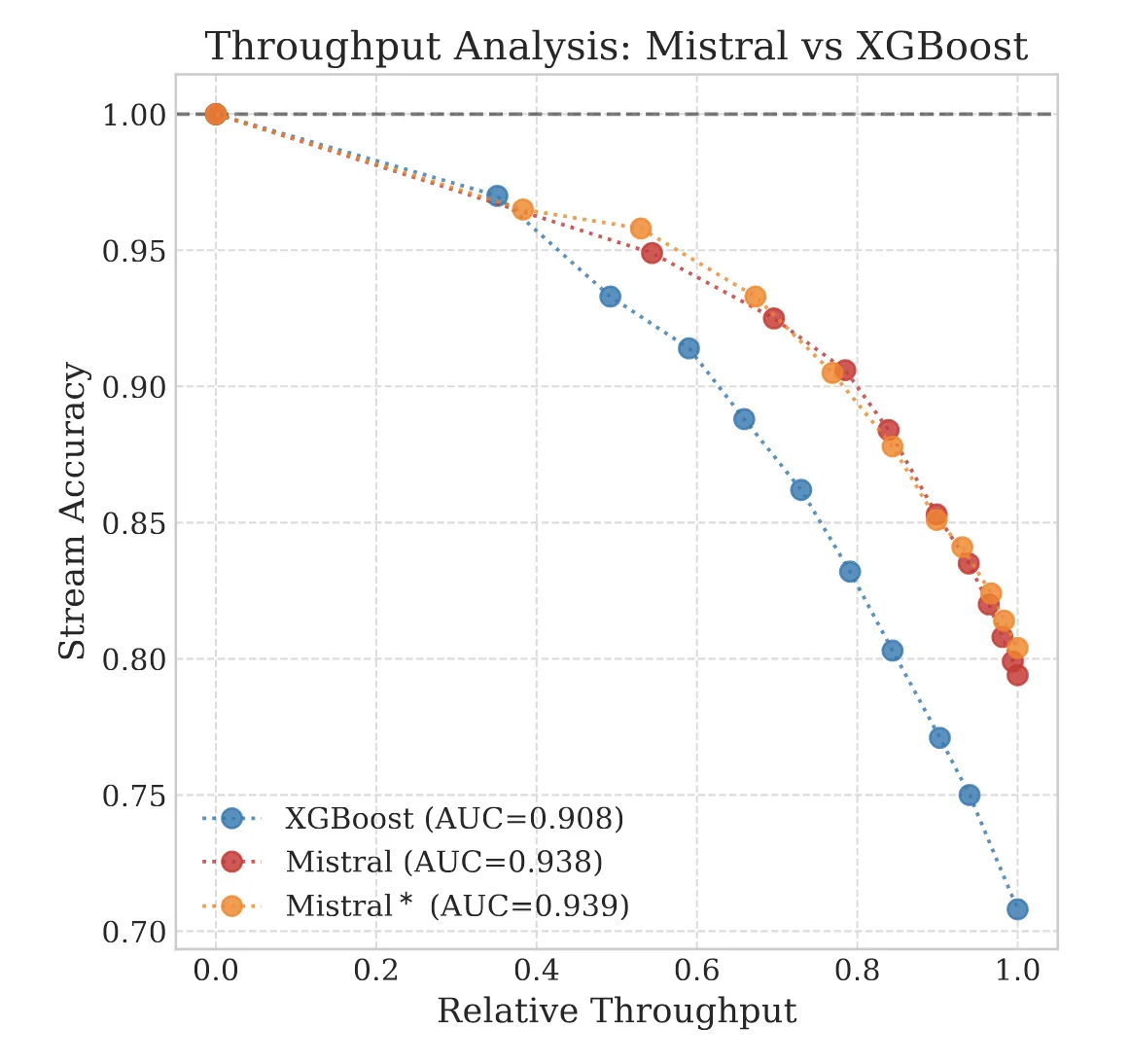
The Reliability Trap: The Limits of 99% Accuracy
We explore the ‘Silent Failure’ mode of LLMs in production: the limits of 99% accuracy for reliability, how confidence decays in long documents, and why standard calibration techniques struggle to fix it.