What kind of paper is this?
This is a Discovery ($\Psi_{\text{Discovery}}$) paper. While it introduces a refined implementation of molecular clock calibration (“cross-bracing”), the primary contribution is the biological inference regarding LUCA’s age, genome size, and metabolic nature. The computational methods serve to characterize a specific biological entity.
What is the motivation?
Understanding the Last Universal Common Ancestor (LUCA) is critical for reconstructing the early evolution of life, yet consensus has been elusive due to disparate data and methods.
- Age Conflicts: Estimates vary widely depending on fossil interpretation and molecular clock calibrations, particularly regarding the “Late Heavy Bombardment” (LHB) constraints.
- Physiological Uncertainty: Debates persist over whether LUCA was a simple “progenote” dependent on geochemistry or a complex prokaryote-grade organism.
- Environmental Context: LUCA is often modeled in isolation, ignoring the ecological interactions that would have shaped its survival and impact on the early Earth system.
What is the novelty here?
The study integrates three advanced computational approaches to provide a holistic reconstruction of LUCA:
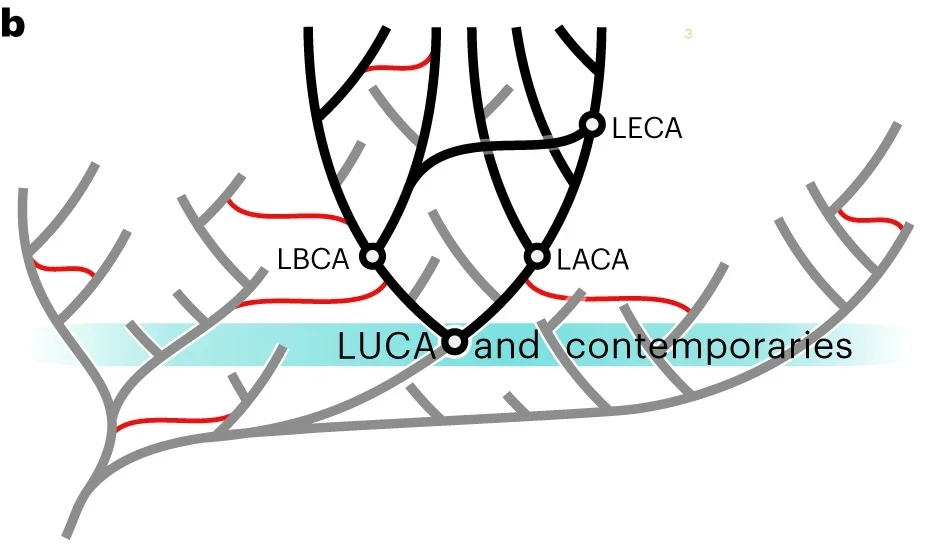
- Cross-Braced Dating: It employs a “cross-bracing” strategy in Bayesian molecular clocks, using pre-LUCA gene duplications (paralogues) to constrain the root. This allows the same fossil calibrations to be applied to mirrored nodes, significantly reducing uncertainty.
- Probabilistic Reconciliation: It uses the ALE (Amalgamated Likelihood Estimation) algorithm to reconcile ~9,300 gene family trees against the species tree. This explicitly models gene transfer, duplication, and loss, allowing for a much broader reconstruction of the proteome.
- Ecosystem Modeling: The physiological reconstruction is coupled with geochemical modeling to propose that LUCA was a member of a productive, hydrogen-recycling early ecosystem.
What experiments were performed?
- Phylogenomics: Inferred a species tree from 57 single-copy marker genes across 700 diverse prokaryotic genomes (350 Archaea, 350 Bacteria) using maximum likelihood (IQ-TREE 2).
- Molecular Dating: Estimated divergence times using MCMCtree with a partitioned dataset of 5 pre-LUCA paralogue pairs (e.g., ATP synthase, EF-Tu/G). Calibrations included 13 fossil constraints and a “soft” maximum bound based on the Moon-forming impact (4.51 Ga).
- Metabolic Reconstruction: Reconciled 9,365 KEGG ortholog families against the species tree to calculate the posterior probability (PP) of each gene’s presence in LUCA. Metabolic potential was inferred from genes with high PP (typically >0.75).
- Genome Size Prediction: Trained a LOESS regression model on modern prokaryotes to predict LUCA’s genome size based on the inferred number of KEGG families.
What outcomes/conclusions?
- Age: LUCA lived approximately 4.2 Ga (95% CI: 4.09-4.33 Ga), surprisingly soon after the Moon-forming impact (~4.5 Ga).
- Complexity: LUCA was a complex, prokaryote-grade organism with a genome size of ~2.75 Mb (encoding ~2,600 proteins), comparable to modern prokaryotes.
- Physiology:
- Metabolism: Anaerobic acetogen (using the Wood-Ljungdahl pathway) capable of fixing $CO_2$ and $N_2$, likely thermophilic (reverse gyrase present).
- Immunity: Possessed an early Class 1 CRISPR-Cas system for antiviral defense.
- Ecology: LUCA likely inhabited an anaerobic environment (hydrothermal vents or surface hot springs) and was part of a community that included methanogens. The study models a global ecosystem where biological $CH_4$ production and atmospheric photochemical recycling of $H_2$ boosted productivity.
Reproducibility Details
Data
The study relied on publicly available genomic data and specific subsets of marker genes.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Phylogeny | Prokaryotic Genomes | 700 genomes | 350 Archaea, 350 Bacteria selected to maximize diversity |
| Dating | Pre-LUCA Paralogues | 5 gene pairs | ATP synthase, Elongation Factor Tu/G, SRP/SRPR, Tyr-tRNA, Leu/Val-tRNA |
| Reconciliation | Gene Families | 9,365 families | Clustered using KEGG Orthology (KO) identifiers |
| Calibration | Fossil/Isotope Records | 13 constraints | Includes max bound at 4.51 Ga (Moon formation) and min bound at 2.95 Ga (oxygenic photosynthesis) |
Algorithms
Key computational steps involved sequence processing, tree inference, and probabilistic reconciliation.
- Alignment & Trimming: sequences aligned with MAFFT L-INS-i (v7.407) and trimmed with BMGE (v1.12, BLOSUM30 matrix, entropy 0.5).
- Tree Inference: IQ-TREE 2 (v2.1.2) using complex mixture models (LG+C60+F+G) to account for site-specific heterogeneity.
- Reconciliation: ALE (Amalgamated Likelihood Estimation) program
ALEml_undatedused to calculate gene presence probabilities, accounting for HGT, duplication, and loss. - Genome Prediction: LOESS regression (Locally Estimated Scatterplot Smoothing) used to map KEGG family counts to total protein counts/genome size.
Models
The analysis employed sophisticated evolutionary models to handle deep time scales and heterogeneity.
- Substitution Models:
- Species Tree:
LG+C60+F+G(mixture model with 60 profiles). - Gene Trees:
LG+C20+F+GorLG+C60+F+Gdepending on alignment properties.
- Species Tree:
- Molecular Clock:
- MCMCtree (PAML v4.10.7).
- Relaxed clock models: GBM (Geometric Brownian Motion) and ILN (Independent Lognormal).
- Cross-Bracing: Specifically models shared divergence times for duplicated nodes (driver and mirror nodes).
Evaluation
Validation focused on robustness across different topologies and clock models.
| Metric | Value | Baseline | Notes |
|---|---|---|---|
| LUCA Age (GBM) | 4.18-4.33 Ga | LHB Hypothesis | Significantly older than LHB constraints often used |
| LUCA Age (ILN) | 4.09-4.32 Ga | - | Consistent across clock models |
| Genome Size | 2.49-2.99 Mb | ~80-1500 genes | Estimates are on the higher end of previous “minimal” gene set theories |
| Topology Test | p > 0.05 | - | AU tests confirmed robustness to uncertainties in CPR and DPANN placement |
Hardware
- Software: PAML v4.10.7 (MCMCtree), IQ-TREE 2, ALE v0.4, HMMER v3.3.2.
- Compute: IQ-TREE runs specified usage of 4 CPUs; MCMCtree approximated likelihood calculation (
approxmethod) to reduce computational cost.
Paper Information
Citation: Moody, E. R. R., Álvarez-Carretero, S., Mahendrarajah, T. A., et al. (2024). The nature of the last universal common ancestor and its impact on the early Earth system. Nature Ecology & Evolution, 8, 1654-1666. https://doi.org/10.1038/s41559-024-02461-1
Publication: Nature Ecology & Evolution 2024
@article{moodyTheNatureLast2024,
title={The nature of the last universal common ancestor and its impact on the early Earth system},
author={Moody, Edmund R. R. and Álvarez-Carretero, Sandra and Mahendrarajah, Tara A. and Clark, James W. and Betts, Holly C. and Dombrowski, Nina and Szánthó, Lénárd L. and Boyle, Richard A. and Daines, Stuart and Chen, Xi and Lane, Nick and Yang, Ziheng and Shields, Graham A. and Szöllősi, Gergely J. and Spang, Anja and Pisani, Davide and Williams, Tom A. and Lenton, Timothy M. and Donoghue, Philip C. J.},
journal={Nature Ecology & Evolution},
volume={8},
number={9},
pages={1654--1666},
year={2024},
publisher={Nature Publishing Group},
doi={10.1038/s41559-024-02461-1}
}
Additional Resources: