What kind of paper is this?
This is a Methodological ($\Psi_{\text{Method}}$) paper. It introduces a novel architecture, InvMSAFold, which hybridizes deep learning encoders with statistical physics-based decoders (Potts models). The rhetorical structure focuses on architectural innovation (low-rank parameter generation), ablation of speed/diversity against baselines (ESM-IF1), and algorithmic efficiency.
What is the motivation?
Standard inverse folding models (like ESM-IF1 or ProteinMPNN) solve a “one-to-one” mapping: given a structure, predict the single native sequence. However, in nature, folding is “many-to-one”: many homologous sequences fold into the same structure.
The authors identify two key gaps:
- Lack of Diversity: Standard autoregressive models maximize probability for the ground truth sequence, often failing to capture the broad evolutionary landscape of viable homologs.
- Slow Inference: Autoregressive sampling requires a full neural network pass for every amino acid, making high-throughput screening (e.g., millions of candidates) computationally prohibitive.
What is the novelty here?
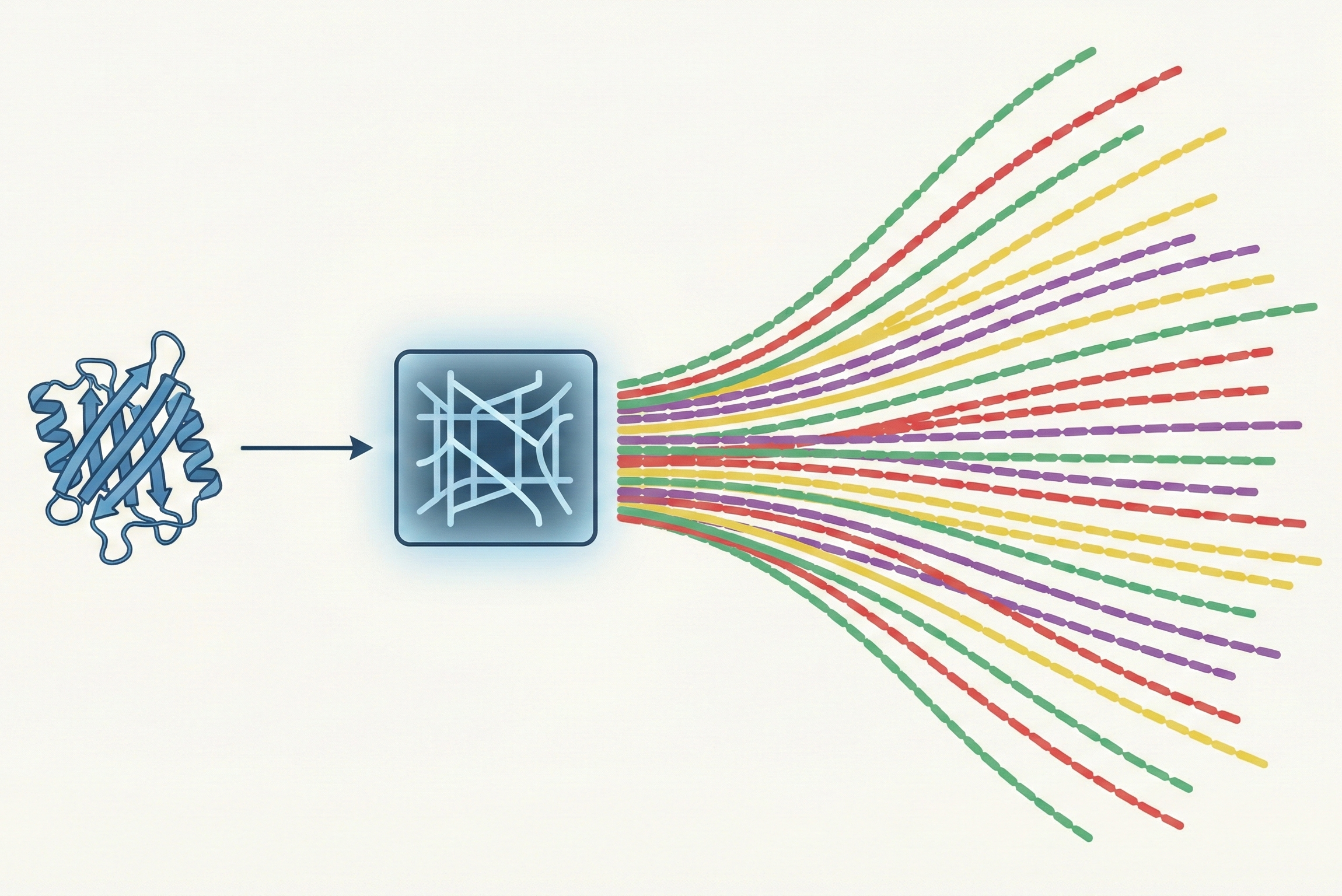
The core novelty is shifting the learning objective from predicting sequences to predicting probability distributions.
InvMSAFold outputs the parameters (couplings $\mathbf{J}$ and fields $\mathbf{h}$) of a Potts Model (a pairwise Markov Random Field).
- Low-Rank Decomposition: To handle the massive parameter space of pairwise couplings ($L xL xq xq$), the model predicts a low-rank approximation $\mathbf{V}$ ($L xK xq$), reducing complexity from $\mathcal{O}(L^2)$ to $\mathcal{O}(L)$.
- One-Shot Generation: The deep network runs only once to generate the Potts parameters. Sampling sequences from this Potts model is then performed on CPU via MCMC or autoregressive approximation, which is orders of magnitude faster than running a Transformer decoder for every step.
What experiments were performed?
The authors validated the model on three CATH-based test sets (Inter-cluster, Intra-cluster, MSA) to test generalization to unseen folds.
- Speed Benchmarking: Compared wall-clock sampling time vs. ESM-IF1 on CPU/GPU.
- Covariance Reconstruction: Checked if generated sequences recover the evolutionary correlations found in natural MSAs (Pearson correlation of covariance matrices).
- Structural Fidelity: Generated sequences with high Hamming distance from native, folded them with AlphaFold 2 (no templates), and measured RMSD to the target structure.
- Property Profiling: Analyzed the distribution of predicted solubility (Protein-Sol) and thermostability (Thermoprot) to demonstrate diversity.
What outcomes/conclusions?
- Massive Speedup: InvMSAFold is orders of magnitude faster than ESM-IF1. Because the “heavy lifting” (generating Potts parameters) happens once, sampling millions of sequences becomes trivial on CPUs.
- Better Diversity: The model captures evolutionary covariances significantly better than ESM-IF1, especially for long sequences.
- Robust Folding: Sequences generated far from the native sequence (high Hamming distance) still fold into the correct structure (low RMSD), whereas ESM-IF1 struggles to produce diverse valid sequences.
- Property Expansion: The method generates a wider spread of biochemical properties (solubility/thermostability), making it more suitable for screening libraries in protein design.
Reproducibility Details
Data
Source: CATH v4.2 database (40% non-redundant dataset).
Splits:
- Training: ~22k domains.
- Inter-cluster Test: 10% of clusters held out (unseen folds).
- Intra-cluster Test: Unseen domains from seen clusters.
- Augmentation: MSAs generated using MMseqs2 against UniRef50. Training uses random subsamples of these MSAs ($|M_X| = 64$) to teach the model evolutionary variance.
Algorithms
Architecture:
- Encoder: Pre-trained ESM-IF1 encoder (frozen or fine-tuned not explicitly specified, likely frozen).
- Decoder: 6-layer Transformer (8 heads) that outputs a latent tensor.
- Projection: Linear layers project latent tensor to fields $\mathbf{h}$ ($L xq$) and low-rank tensor $\mathbf{V}$ ($L xK xq$).
Coupling Construction: The full coupling tensor $\mathcal{J}$ is approximated via: $$\mathcal{J}_{i,a,j,b} = \frac{1}{\sqrt{K}} \sum_{k=1}^{K} \mathcal{V}_{i,k,a} \mathcal{V}_{j,k,b}$$ Rank $K=48$ was used.
Loss Functions: Two variants were trained:
- InvMSAFold-PW: Trained via Pseudo-Likelihood (PL). Computation is optimized to $\mathcal{O}(L)$ time using the low-rank property.
- InvMSAFold-AR: Trained via Autoregressive Likelihood. Couplings are masked ($J_{ij} = 0$ if $i<j$) to allow exact likelihood computation and direct sampling without MCMC.
Models
- InvMSAFold-PW: Requires MCMC sampling (Metropolis-Hastings) at inference.
- InvMSAFold-AR: Allows direct, fast autoregressive sampling.
- Hyperparameters: AdamW optimizer, lr=$10^{-4}$ (PW) / $3.4 x10^{-4}$ (AR), 94 epochs. L2 regularization on fields/couplings.
Evaluation
Metrics:
- scRMSD: Structure fidelity (AlphaFold2 prediction vs. Native).
- Covariance Pearson Correlation: Measures recovery of evolutionary pairwise statistics.
- Sampling Speed: Wall-clock time vs. sequence length/batch size.
Hardware
- Training: Not specified, but “fast” due to low rank.
- Inference:
- ESM-IF1: NVIDIA GeForce RTX 4060 Laptop (8GB).
- InvMSAFold: Single core of Intel i9-13905H CPU.
Citation
@inproceedings{silvaFastUncoveringProtein2024,
title = {Fast {{Uncovering}} of {{Protein Sequence Diversity}} from {{Structure}}},
booktitle = {The {{Thirteenth International Conference}} on {{Learning Representations}}},
author = {Silva, Luca Alessandro and {Meynard-Piganeau}, Barthelemy and Lucibello, Carlo and Feinauer, Christoph},
year = 2024,
month = oct,
url = {https://openreview.net/forum?id=1iuaxjssVp}
}