What kind of paper is this?
This is primarily a Theory paper ($\Psi_{\text{Theory}}$) with a strong Systematization component ($\Psi_{\text{Systematization}}$).
- Theory: It applies statistical mechanics (specifically spin glass theory) to derive formal relationships between energy barriers, entropy, and folding kinetics.
- Systematization: It synthesizes two previously conflicting views (specific “folding pathways” versus thermodynamic “funnels”) into a unified phase diagram.
What is the motivation?
The work addresses Levinthal’s Paradox: the disconnect between the astronomical number of possible conformations (requiring $10^{10}$ years to search randomly) and the millisecond-to-second timescales observed in biology.
- The Conflict: Previous theories often relied on specific, unique folding pathways (a concept Levinthal originally proposed to resolve his own paradox) or distinct intermediates. The authors argue these are insufficient to explain the robustness of folding.
- The Gap: There was a need to quantitatively distinguish between sequences that fold reliably (“good folders”) and random heteropolymers that get trapped in local minima (glassy states).
What is the novelty here?
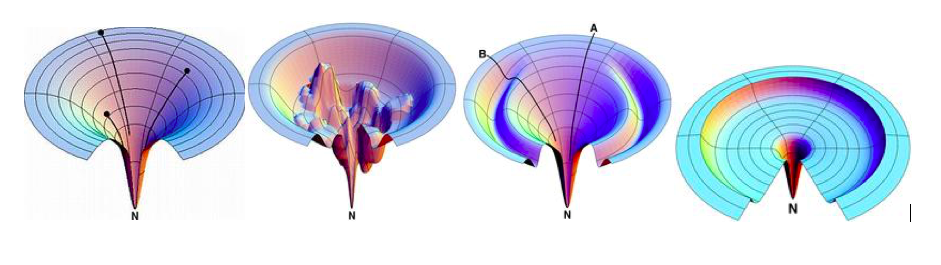
The core novelty is the Energy Landscape Theory, which posits that proteins fold via a “funnel”.
- Folding Funnel & Reaction Coordinate ($n$): The landscape is defined over a reaction coordinate $n$, representing structural similarity to the native state ($n=1$ is native, $n=0$ is unfolded). The funnel drives the protein from high-entropy, high-energy states (low $n$) to the low-entropy, low-energy native state (high $n$).
- Kinetic vs. Thermodynamic Bottlenecks: A crucial departure from classical transition state theory is the distinction between the thermodynamic bottleneck ($n^\dagger_{th}$, where free energy is highest) and the kinetic bottleneck ($n^\dagger_{kin}$, where the folding flow is most restricted). These do not always coincide, meaning the rate-limiting step can shift with temperature.
- Principle of Minimal Frustration: Natural proteins are evolved to minimize conflicting interactions. This frustration comes in two forms: energetic (competing favorable interactions) and topological/geometric (steric hindrances). Minimizing these creates a smooth funnel.
- The “Ferry Law” (Escape Time): The theory provides a rigorous “super-Arrhenius” law for the time required to escape local traps in a rough landscape: $$ \tau(n) = \tau_0 \exp\left[ \left( \frac{\Delta E(n)}{k_B T} \right)^2 \right] $$ This highlights how landscape roughness ($\Delta E$) drastically increases folding time as temperature decreases.
- Stability Gap: The energy gap ($\Delta E_s$) separating the native state from the glass transition. Maximizing this gap (relative to the glass transition temperature, $T_f / T_g$) ensures foldability.
- Folding Scenarios: The definition of distinct kinetic scenarios based on the relationship between the glass transition location ($n_g$) and the thermodynamic bottleneck ($n^\dagger$).
Scenario Characteristics Kinetics Type 0A Downhill folding No barrier; $n^\dagger = 0$. Extremely fast. Type 0B Downhill folding with glass Glass transition occurs, but no thermodynamic barrier. Type I Two-state folding ($T_f > T_g$) Standard barrier crossing; $n_g$ is irrelevant or far from $n^\dagger$. Self-averaging, smooth exponential kinetics. Type IIA Glassy folding ($n^\dagger < n_g$) Glass transition occurs after the bottleneck. Kinetics are mostly single-exponential but can trap late. Type IIB Glassy folding ($n^\dagger \ge n_g$) Glass transition occurs before or at the bottleneck. Non-self-averaging; kinetics depend strictly on sequence details.
What experiments were performed?
The authors performed analytical derivations and lattice simulations to validate the theory.
- Lattice Simulations: They simulated 27-mer heteropolymers on a cubic lattice using Monte Carlo methods.
- Sequence Variation: They compared “designed” sequences (unfrustrated) against random sequences to observe differences in collapse and folding times.
- Phase Diagram Mapping: They mapped the behavior of these polymers onto a Phase Diagram (Temperature vs. Landscape Roughness $\Delta E$), predicting regions of random coil, globule, folded, and glass states.
What outcomes/conclusions?
- Folding is Ensemble-Based: Folding involves the simultaneous “funneling” of an ensemble of conformations toward the native state.
- Self-Averaging vs. Non-Self-Averaging:
- Self-Averaging: Properties depend only on the overall composition (e.g., hydrophobic/polar ratio), meaning mutations have little effect.
- Non-Self-Averaging: In the glassy phase ($T < T_g$), folding kinetics depend strictly on the detailed sequence; single mutations can drastically alter pathways.
- Curved Arrhenius Plots: The theory predicts curved (parabolic) Arrhenius plots due to the location of the kinetic bottleneck shifting with temperature and landscape roughness. Note that in experimental settings, this curvature is often ascribed to the temperature dependence of the hydrophobic effect ($\Delta C_p$), a distinct mechanism from the model’s bottleneck shift.
- Optimization Criterion: To engineer fast-folding proteins, one must maximize the stability gap ratio ($T_f/T_g$).
- Experimental Validation: The authors map real-world proteins to the theoretical scenarios: Chymotrypsin Inhibitor 2 (CI2) behaves as a Type I folder (two-state), Hen Lysozyme maps to Type II (glassy intermediates at high temperature), and Cytochrome c fits the Type 0 scenario (downhill folding).
Reproducibility Details
The simulations are based on the “27-mer” cubic lattice model, a standard paradigm in theoretical protein folding.
Data
The “data” consists of specific synthetic sequences used in the Monte Carlo simulations.
| Sequence ID | Sequence (27-mer) | Type | $T_f$ |
|---|---|---|---|
| 002 | ABABBBBBABBABABAAABBAAAAAAB | Optimized | 1.285 |
| 004 | AABAABAABBABAAABABBABABABBB | Optimized | 1.26 |
| 006 | AABABBABAABBABAAAABABAABBBB | Random | 0.95 |
| 013 | ABBBABBABAABBBAAABBABAABABA | Random | 0.83 |
- Source: Table I in the paper.
- Alphabet: Two-letter code (A/B), representing hydrophobic/polar distinctions.
Algorithms
- Simulation Method: Monte Carlo (MC) sampling on a discrete lattice.
- Glass Transition ($T_g$) Definition: Defined kinetically where the folding time $\tau_f(T_g)$ exceeds $(\tau_{max} + \tau_{min})/2$. In this study, $\tau_{max} = 1.08 \times 10^9$ MC steps.
- Folding Temperature ($T_f$): Calculated using the Monte Carlo histogram method, defined as the temperature where the probability of occupying the native structure is 0.5.
Models
- Lattice: 27 monomers on a $3 \times 3 \times 3$ cubic lattice (maximally compact states can be fully enumerated).
- Potential Energy:
- Interactions occur between nearest neighbors on the lattice that are not covalently connected.
- $E_{AA} = E_{BB} = -3$ (Strong attraction for like pairs).
- $E_{AB} = -1$ (Weak attraction for unlike pairs).1
- Frustration: Defined via the $Q$ measure (similarity to ground state). “Frustrated” sequences have low-energy states that are structurally dissimilar (low $Q$) to the ground state.
Evaluation
- Folding Time ($\tau$): Mean first passage time (MFPT) to reach the native structure from a random coil.
- Collapse Time: Time required to reach a conformation with 25 or 28 contacts for the first time.
- Reaction Coordinate: The similarity measure $n$ (or $Q$), typically defined as the number of native contacts formed.
Paper Information
Citation: Bryngelson, J. D., Onuchic, J. N., Socci, N. D., & Wolynes, P. G. (1995). Funnels, Pathways, and the Energy Landscape of Protein Folding: A Synthesis. Proteins: Structure, Function, and Genetics, 21(3), 167-195. https://doi.org/10.1002/prot.340210302
Publication: Proteins 1995
@article{bryngelson1995funnels,
title={Funnels, Pathways, and the Energy Landscape of Protein Folding: A Synthesis},
author={Bryngelson, Joseph D. and Onuchic, José Nelson and Socci, Nicholas D. and Wolynes, Peter G.},
journal={Proteins: Structure, Function, and Genetics},
volume={21},
number={3},
pages={167--195},
year={1995},
doi={10.1002/prot.340210302}
}
Additional Resources:
Note: The main text of the original paper mentions an interaction of +1 for unlike pairs, but the Figure 2 caption and subsequent usage imply -1. We assume -1 for consistency with standard attractive potentials. ↩︎