What kind of paper is this?
This is primarily a Perspective paper (with Theory and Discovery elements):
- Perspective: Defines a “Grand Challenge” and argues for a conceptual shift in how we view biomolecular assembly
- Theory: Uses formal combinatorial arguments to establish the bounds of the search space ($10^{300}$ configurations)
- Discovery: Uses experimental data on alkaline phosphatase to validate the kinetic hypothesis
What is the motivation?
The Central Question: How does a protein choose one unique structure out of a hyper-astronomical number of possibilities in a biological timeframe (seconds)?
Levinthal provides a “back-of-the-envelope” derivation to define the problem scope:
- Degrees of Freedom: A generic, unrestricted protein with 2,000 atoms would possess ~6,000 degrees of freedom. However, physical constraints (specifically the planar peptide bond) reduce this significantly. For a 150-amino acid protein, these constraints lower the complexity to ~450 degrees of freedom (300 rotations, 150 bond angles).
- The Combinatorial Explosion: Even with conservative estimates, this results in $10^{300}$ possible conformations.
- The Time Constraint: Since proteins fold in seconds, Levinthal calculates they can sample at most $10^8$ conformations (based on atomic vibration times) before stabilizing. Against $10^{300}$ possibilities, this search effectively covers 0% of the space, proving the impossibility of random search.
The Insight: The existence of folded proteins proves the impossibility of random global search. The system must be guided.
What is the novelty here?
Core Contribution: Levinthal reframes folding from a thermodynamic problem (seeking the absolute global minimum) to a Kinetic Control problem. He argues the native state is a “metastable” energy well found quickly by a specific pathway, which can differ from the system’s lowest possible energy state.
The Pathway Dependence Hypothesis
The key insights of kinetic control:
- Nucleation: The process is “speeded and guided by the rapid formation of local interactions”
- Pathway Constraints: These local interactions (likely within proximal amino acids) serve as nucleation points that restrict further folding, effectively pruning the search space
- The “Metastable” State: The final structure represents a “metastable state” in a sufficiently deep energy well that is kinetically accessible via the folding pathway, independent of the global energy minimum.
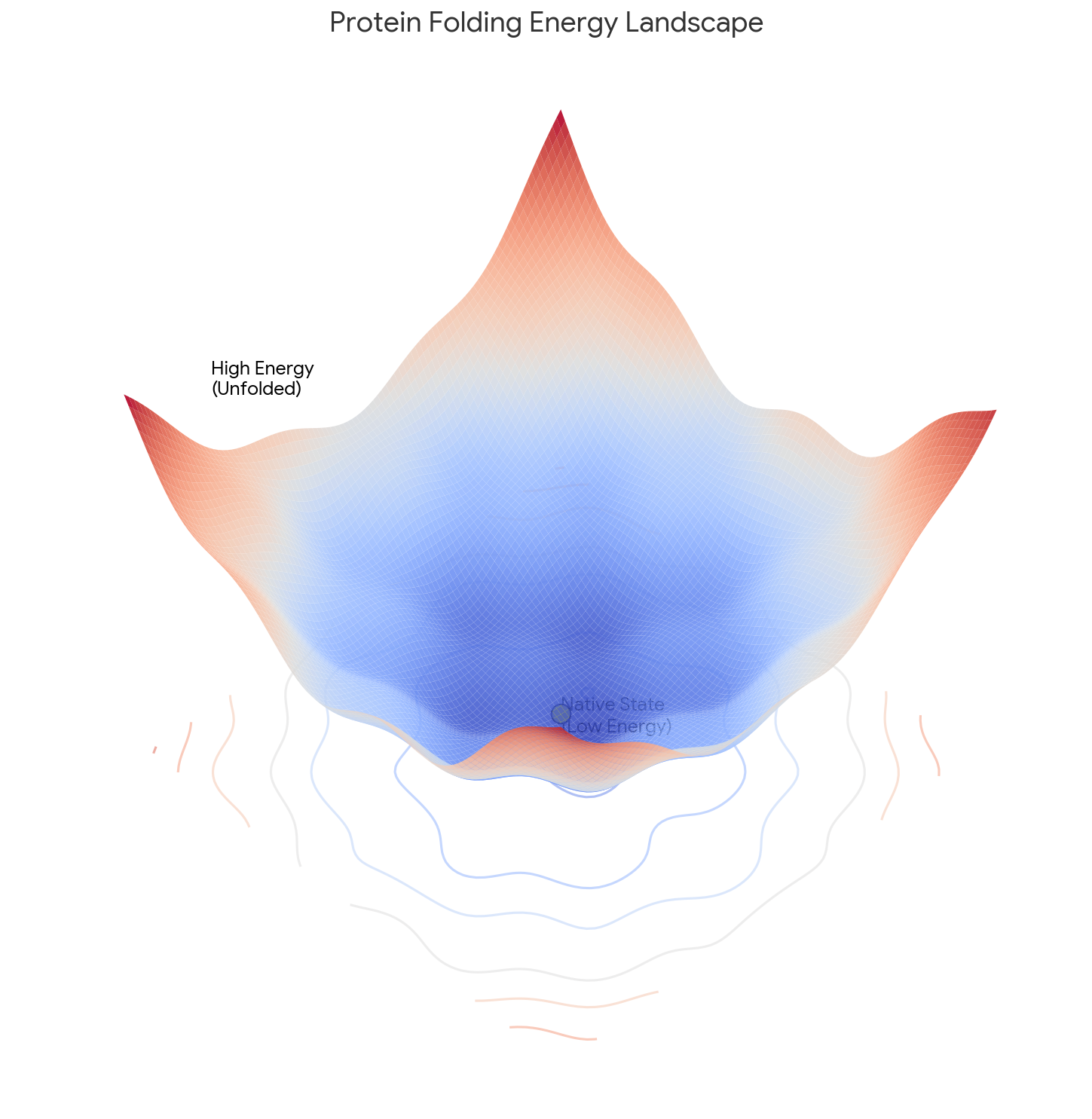
What experiments were performed?
To support the pathway hypothesis, Levinthal cites work on Alkaline Phosphatase (MW ~40,000), utilizing its property as a dimer of two identical subunits:
- Renaturation Window: The wild-type enzyme refolds optimally at 37°C. However, mutants were isolated that only produce active enzyme (and renature) at temperatures below 37°C.
- Stability vs. Formation: Crucially, once folded, both the wild-type and mutant enzymes are stable up to 90°C.
- The Rate-Limiting Step: Levinthal notes that the rate-limiting step for activity is the formation of the dimer from monomers. This proves that the order of assembly (kinetic pathway) dictates the final structure, distinct from the final structure’s thermodynamic stability.
What outcomes/conclusions?
Key Finding
The mutant experiments serve as the “smoking gun”: a protein seeking a global thermodynamic minimum would fold spontaneously at any temperature where the final state is stable (up to 90°C). The fact that mutants require specific lower temperatures for formation (while remaining stable at high temperatures once formed) proves that the kinetic pathway determines the outcome alongside the thermodynamic endpoint.
Broader Implications
Levinthal explicitly asks: “Is a unique folding necessary for any random 150-amino acid sequence?” and answers “Probably not.” This anticipates that valid proteins lie on a low-dimensional manifold within sequence space; most random sequences remain unfolded.
He concludes by connecting these computational models to Mössbauer spectroscopy, noting that physical simulations are essential to understand how small structural fluctuations in polypeptides affect nuclear resonance signals (a reminder of the specific conference context where this perspective was delivered).
Connection to Modern AI for Science
Understanding this paper is crucial for reading modern deep learning papers on protein folding (e.g., AlphaFold, ESMFold):
- From Oscilloscopes to GPUs: Levinthal actively simulated folding on 1960s hardware, using computer-controlled oscilloscopes and vector matrix multiplications to visualize 3D protein structures and motion pictures. This pioneered the field of in silico folding that now runs on massive GPU clusters.
- Inductive Bias: Levinthal’s observation that “local interactions” guide folding provides the biological justification for Graph Neural Networks (GNNs) and Attention mechanisms that prioritize local neighbor connectivity ($k$-nearest neighbors) in molecular representation learning
- The Optimization Landscape: Levinthal’s rejection of “random search” prefigures the need for modern optimization techniques (like Gradient Descent) that navigate high-dimensional landscapes by following local gradients.
- Generative AI & The Manifold Hypothesis: The insight that most random sequences remain unfolded underpins the core challenge of Inverse Folding (Protein Design). Generative models (like ProteinMPNN) optimize the boundaries of the “foldable” space that Levinthal first defined.
Paper Information
Citation: Levinthal, C. (1969). How to Fold Graciously. In Mössbauer Spectroscopy in Biological Systems: Proceedings of a meeting held at Allerton House, Monticello, Illinois (pp. 22-24). University of Illinois Press.
Publication: Mössbauer Spectroscopy in Biological Systems Proceedings, 1969
@inproceedings{levinthal1969fold,
title={How to fold graciously},
author={Levinthal, Cyrus},
booktitle={M{\"o}ssbauer spectroscopy in biological systems},
pages={22--24},
year={1969},
organization={University of Illinois Press Urbana},
url={https://faculty.cc.gatech.edu/~turk/bio_sim/articles/proteins_levinthal_1969.pdf}
}
Additional Resources:
