Key Contribution
MARCEL contributes a large-scale dataset for molecular representation and conformer ensemble learning, facilitating advancements in drug discovery and cheminformatics.
| Dataset Details | |
| Authors | Yanqiao Zhu, Jeehyun Hwang, Brock Anton Stenfors, Yuanqi Du, Olexandr Isayev, Keir Adams, Jatin Chauhan, Connor W. Coley, Yizhou Sun, Zhen Liu, Bozhao Nan, Olaf Wiest, Wei Wang |
| Paper Title | Learning over Molecular Conformer Ensembles: Datasets and Benchmarks |
| Institutions | UCLA, MIT, CMU, Notre Dame, Cornell |
| Published In | International Conference on Learning Representations |
| Category | Computational Chemistry |
| Format | SMILES RDKit mol objects 3D coordinates Statistical weights Experimental properties |
| Size | Conformers: 722,193 Molecules: 76,651 Reactions: 6,787 |
| Date | September 2025 |
| Year | 2024 |
| Links | 📊 Dataset • 📄 Paper |
MARCEL contributes a large-scale dataset for molecular representation and conformer ensemble learning, facilitating advancements in drug discovery and cheminformatics.
| Type | Count |
|---|---|
| Conformers | 722,193 |
| Molecules | 76,651 |
| Reactions | 6,787 |
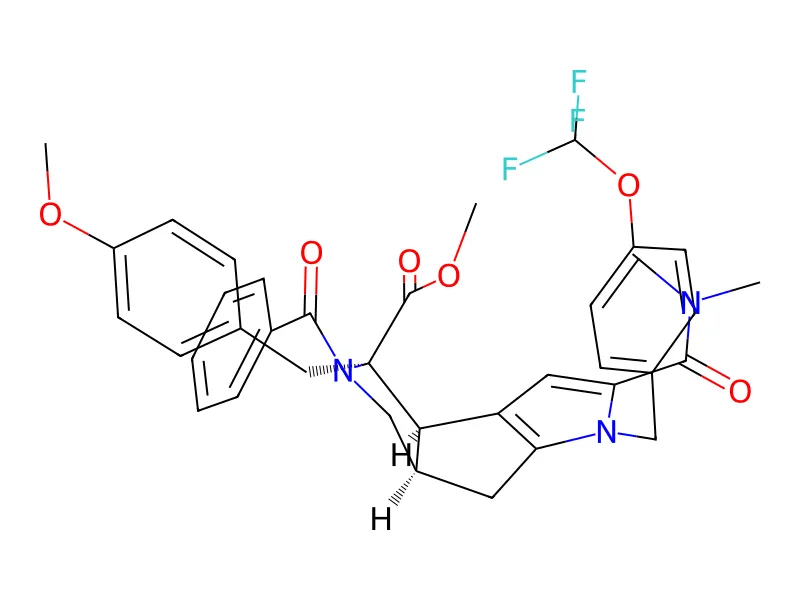
![Example conformer from Drugs-75K (SMILES: COC(=O)[C@@]1(Cc2ccc(OC)cc2)[C@H]2c3cc(C(=O)N(C)C)n(Cc4ccc(OC(F)(F)F)cc4)c3C[C@H]2CN1C(=O)c1ccccc1; IUPAC: methyl (2R,3R,6R)-4-benzoyl-10-(dimethylcarbamoyl)-3-[(4-methoxyphenyl)methyl]-9-[[4-(trifluoromethoxy)phenyl]methyl]-4,9-diazatricyclo[6.3.0.02,6]undeca-1(8),10-diene-3-carboxylate)](/img/marcel-drugs-75k-example-conformer.gif)

| Subset | Count | Description |
|---|---|---|
| Drugs-75K | 75,099 | Drug-like molecules with at least 5 rotatable bonds |
| Kraken | 1,552 | monodentate organophosphorus (III) ligands |
| EE | 872 | Rhodium (Rh)-bound atropisomeric catalysts derived from chiral bisphosphine |
| BDE | 5,195 | Organometallic catalysts ML$_1$L$_2$ |
| Model | IP | EA | χ |
|---|---|---|---|
| 1D - Random forest | 0.4987 | 0.4747 | 0.2732 |
| 1D - LSTM | 0.4788 | 0.4648 | 0.2505 |
| 1D - Transformer | 0.6617 | 0.5850 | 0.4073 |
| 2D - GIN | 0.4354 | 0.4169 | 0.2260 |
| 2D - GIN+VN | 0.4361 | 0.4169 | 0.2267 |
| 2D - ChemProp | 0.4595 | 0.4417 | 0.2441 |
| 2D - GraphGPS | 0.4351 | 0.4085 | 0.2212 |
| 3D - SchNet | 0.4394 | 0.4207 | 0.2243 |
| 3D - DimeNet++ | 0.4441 | 0.4233 | 0.2436 |
| 3D - GemNet | 🥈 0.4069 | 🥈 0.3922 | 🥇 0.1970 |
| 3D - PaiNN | 0.4505 | 0.4495 | 0.2324 |
| 3D - ClofNet | 0.4393 | 0.4251 | 0.2378 |
| 3D - LEFTNet | 0.4174 | 0.3964 | 0.2083 |
| Ensemble - SchNet | 0.4452 | 0.4232 | 0.2243 |
| Ensemble - DimeNet++ | 0.4126 | 0.3944 | 0.2267 |
| Ensemble - GemNet | 🥇 0.4066 | 🥇 0.3910 | 🥈 0.2027 |
| Ensemble - PaiNN | 0.4466 | 0.4269 | 0.2294 |
| Ensemble - ClofNet | 0.4280 | 0.4033 | 0.2199 |
| Ensemble - LEFTNet | 0.4149 | 0.3953 | 0.2069 |
| Model | B₅ | L | BurB₅ | BurL |
|---|---|---|---|---|
| 1D - Random forest | 0.4760 | 0.4303 | 0.2758 | 0.1521 |
| 1D - LSTM | 0.4879 | 0.5142 | 0.2813 | 0.1924 |
| 1D - Transformer | 0.9611 | 0.8389 | 0.4929 | 0.2781 |
| 2D - GIN | 0.3128 | 0.4003 | 0.1719 | 0.1200 |
| 2D - GIN+VN | 0.3567 | 0.4344 | 0.2422 | 0.1741 |
| 2D - ChemProp | 0.4850 | 0.5452 | 0.3002 | 0.1948 |
| 2D - GraphGPS | 0.3450 | 0.4363 | 0.2066 | 0.1500 |
| 3D - SchNet | 0.3293 | 0.5458 | 0.2295 | 0.1861 |
| 3D - DimeNet++ | 0.3510 | 0.4174 | 0.2097 | 0.1526 |
| 3D - GemNet | 0.2789 | 0.3754 | 0.1782 | 0.1635 |
| 3D - PaiNN | 0.3443 | 0.4471 | 0.2395 | 0.1673 |
| 3D - ClofNet | 0.4873 | 0.6417 | 0.2884 | 0.2529 |
| 3D - LEFTNet | 0.3072 | 0.4493 | 0.2176 | 0.1486 |
| Ensemble - SchNet | 0.2704 | 0.4322 | 0.2024 | 0.1443 |
| Ensemble - DimeNet++ | 0.2630 | 🥈 0.3468 | 0.1783 | 🥈 0.1185 |
| Ensemble - GemNet | 🥈 0.2313 | 🥇 0.3386 | 🥇 0.1589 | 🥇 0.0947 |
| Ensemble - PaiNN | 🥇 0.2225 | 0.3619 | 🥈 0.1693 | 0.1324 |
| Ensemble - ClofNet | 0.3228 | 0.4485 | 0.2178 | 0.1548 |
| Ensemble - LEFTNet | 0.2644 | 0.3643 | 0.2017 | 0.1386 |
| Model | EE | BDE |
|---|---|---|
| 1D - Random forest | 61.2963 | 3.0335 |
| 1D - LSTM | 64.0088 | 2.8279 |
| 1D - Transformer | 62.0816 | 10.0771 |
| 2D - GIN | 62.3065 | 2.6368 |
| 2D - GIN+VN | 62.3815 | 2.7417 |
| 2D - ChemProp | 61.0336 | 2.6616 |
| 2D - GraphGPS | 61.6251 | 2.4827 |
| 3D - SchNet | 17.7421 | 2.5488 |
| 3D - DimeNet++ | 14.6414 | 🥇 1.4503 |
| 3D - GemNet | 18.0338 | 1.6530 |
| 3D - PaiNN | 20.2359 | 2.1261 |
| 3D - ClofNet | 33.9473 | 2.6057 |
| 3D - LEFTNet | 19.7974 | 🥈 1.5328 |
| Ensemble - SchNet | 14.2238 | 1.9737 |
| Ensemble - DimeNet++ | 🥈 12.0259 | 1.4741 |
| Ensemble - GemNet | 🥇 11.6142 | 1.6059 |
| Ensemble - PaiNN | 13.5570 | 1.8744 |
| Ensemble - ClofNet | 13.9647 | 2.0106 |
| Ensemble - LEFTNet | 18.4189 | 1.5276 |
Uses the original dataset directly, but only focuses on 5 of the 78 properties.
| Dataset | Relationship | Link |
|---|---|---|
| GEOM | Contains Subset | 📄 View Details |
| Kraken | Contains | N/A |
| EE | Contains | N/A |
| BDE | Contains | N/A |