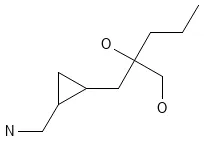
Key Contribution
The main contribution is the creation and release of the 977.4 million-compound GDB-13, a massive expansion in molecular size (up to 13 atoms) and elemental diversity (including S and Cl) made possible by key algorithmic optimizations that significantly accelerated the enumeration process.