Paper Information
Citation: Chang, Q., Chen, M., Pi, C., et al. (2024). RFL: Simplifying Chemical Structure Recognition with Ring-Free Language. arXiv preprint arXiv:2412.07594. https://doi.org/10.48550/arXiv.2412.07594
Publication: arXiv preprint (December 2024)
Additional Resources:
What kind of paper is this?
This is a Methodological paper ($\Psi_{\text{Method}}$). It introduces a novel representation system (Ring-Free Language) and a specialized neural architecture (Molecular Skeleton Decoder) designed to solve specific limitations in converting 2D images to 1D chemical strings. The paper validates this method through direct comparison with state-of-the-art baselines and ablation studies.
What is the motivation?
Current Optical Chemical Structure Recognition (OCSR) methods typically rely on “unstructured modeling,” where 2D molecular graphs are flattened into 1D strings like SMILES or SSML. While simple, these linear formats struggle to explicitly capture complex spatial relationships, particularly in molecules with multiple rings and branches. End-to-end models often fail to “understand” the graph structure when forced to predict these implicit 1D sequences, leading to error accumulation in complex scenarios.
What is the novelty here?
The authors propose two primary contributions to decouple spatial complexity:
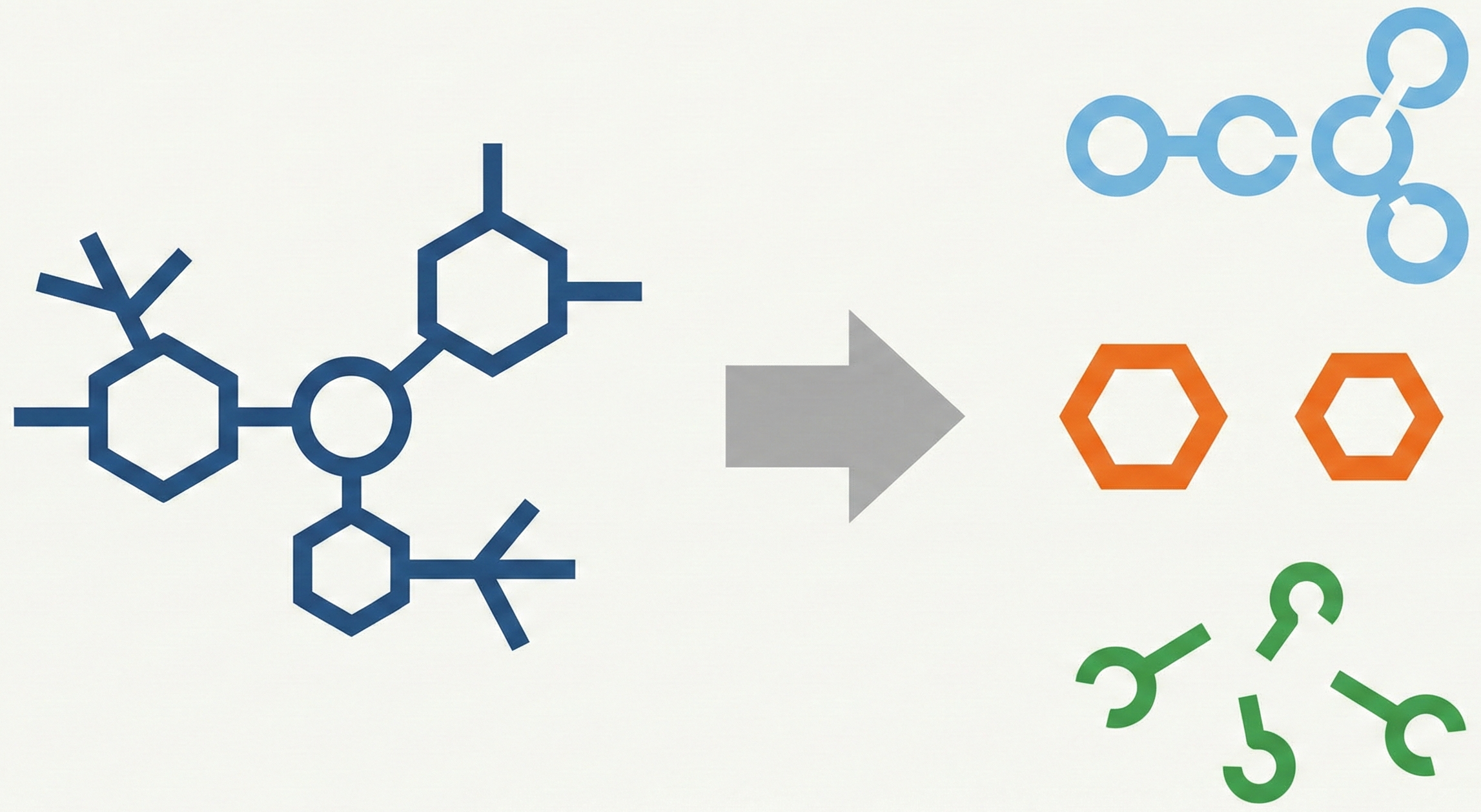
- Ring-Free Language (RFL): A divide-and-conquer representation that splits a molecular graph $G$ into three explicit components: a molecular skeleton $\mathcal{S}$, individual ring structures $\mathcal{R}$, and branch information $\mathcal{F}$. This allows rings to be collapsed into “SuperAtoms” or “SuperBonds” during initial parsing.
- Molecular Skeleton Decoder (MSD): A hierarchical architecture that progressively predicts the skeleton first, then the individual rings (using SuperAtom features as conditions), and finally classifies the branch connections.
What experiments were performed?
The method was evaluated on both handwritten and printed chemical structures against two baselines: DenseWAP (Zhang et al. 2018) and RCGD (Hu et al. 2023).
- Datasets:
- EDU-CHEMC: ~49k handwritten samples (challenging, diverse styles)
- Mini-CASIA-CSDB: ~89k printed samples (from ChEMBL)
- Synthetic Complexity Dataset: A custom split of ChEMBL data grouped by structural complexity (atoms + bonds + rings) to test generalization
- Ablation Studies: Tested the necessity of the MSD architecture vs. standard decoders and the impact of the
[conn]token for filtering branch candidates
What were the outcomes and conclusions drawn?
- SOTA Performance: The proposed method (MSD-RCGD) outperformed the previous state-of-the-art (RCGD) on both handwritten (95.38% Exact Match) and printed (95.58% Exact Match) datasets.
- Universal Improvement: Applying MSD/RFL to an older baseline (DenseWAP) improved its accuracy significantly (e.g., from 87.6% to 81.9% on complex sets), proving the method is model-agnostic.
- Complexity Handling: The method showed robust generalization on unseen high-complexity molecules, where standard 1D baselines failed completely (0% accuracy for standard DenseWAP vs. ~30% for MSD version on the hardest tier).
Reproducibility Details
Data
The authors utilized one handwritten and one printed dataset, plus a synthetic set for stress-testing complexity.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Training/Test | EDU-CHEMC | 48,998 Train / 2,992 Test | Handwritten images from educational scenarios |
| Training/Test | Mini-CASIA-CSDB | 89,023 Train / 8,287 Test | Printed images rendered from ChEMBL using RDKit |
| Generalization | ChEMBL Subset | 5 levels of complexity | Custom split based on Eq: $N_{atom} + N_{bond} + 12 \xN_{ring}$ |
Algorithms
RFL Splitting (Encoding):
- Detect Rings: Use DFS to find all non-nested rings $\mathcal{R}$.
- Determine Adjacency ($\gamma$): Calculate shared edges between rings.
- Merge:
- If $\gamma(r_i) = 0$ (isolated), merge ring into a SuperAtom node.
- If $\gamma(r_i) > 0$ (adjacent), merge ring into a SuperBond edge.
- Update: Record connection info in $\mathcal{F}$ and remove ring details from the main graph to form Skeleton $\mathcal{S}$.
MSD Decoding:
- Hierarchical Prediction: The model predicts the Skeleton $\mathcal{S}$ first.
- Contextual Ring Prediction: When a SuperAtom/Bond token is predicted, its hidden state $f^s$ is stored. After the skeleton is finished, $f^s$ is used as a condition to autoregressively decode the specific ring structure.
- Token
[conn]: A special token separates connected ring bonds from unconnected ones to sparsify the branch classification task.
Models
The architecture follows a standard Image-to-Sequence pattern but with a forked decoder.
- Encoder: DenseNet (Growth rate=24, Depth=32 per block)
- Decoder (MSD):
- Core: GRU with Attention (Hidden dim=256, Embedding dim=256, Dropout=0.15)
- Skeleton Module: Autoregressively predicts sequence tokens. Uses Maxout activation.
- Branch Module: A binary classifier (MLP) taking concatenated features of skeleton bonds $f_{bs}$ and ring bonds $f_{br}$ to predict connectivity matrix $\mathcal{F}$.
- Loss Function: $\mathcal{O} = \lambda_1 \mathcal{L}{ce} + \lambda_2 \mathcal{L}{cls}$ (where $\lambda=1$)
Evaluation
Metrics focus on exact image reconstruction and structural validity.
| Metric | Description | Notes |
|---|---|---|
| EM (Exact Match) | % of images where predicted graph exactly matches ground truth. | Primary metric |
| Struct-EM | % of correctly identified chemical structures (ignoring non-chemical text). | Auxiliary metric |
Hardware
- Compute: 4 x NVIDIA Tesla V100 (32GB RAM)
- Training Configuration:
- Batch size: 8 (Handwritten), 32 (Printed)
- Epochs: 50
- Optimizer: Adam ($lr=2\times10^{-4}$, decayed by 0.5 via MultiStepLR)
Citation
@misc{changRFLSimplifyingChemical2025,
title = {RFL: Simplifying Chemical Structure Recognition with Ring-Free Language},
shorttitle = {RFL},
author = {Chang, Qikai and Chen, Mingjun and Pi, Changpeng and Hu, Pengfei and Zhang, Zhenrong and Ma, Jiefeng and Du, Jun and Yin, Baocai and Hu, Jinshui},
year = {2024},
month = dec,
number = {arXiv:2412.07594},
eprint = {2412.07594},
primaryclass = {cs},
publisher = {arXiv},
doi = {10.48550/arXiv.2412.07594},
archiveprefix = {arXiv}
}