Paper Information
Citation: Fan, S., Xie, Y., Cai, B., Xie, A., Liu, G., Qiao, M., Xing, J., & Nie, Z. (2025). OCSU: Optical Chemical Structure Understanding for Molecule-centric Scientific Discovery. arXiv preprint arXiv:2501.15415. https://doi.org/10.48550/arXiv.2501.15415
Publication: arXiv 2025
Additional Resources:
What kind of paper is this?
This is primarily a Methodological Paper ($\Psi_{\text{Method}}$) with a significant Resource ($\Psi_{\text{Resource}}$) contribution.
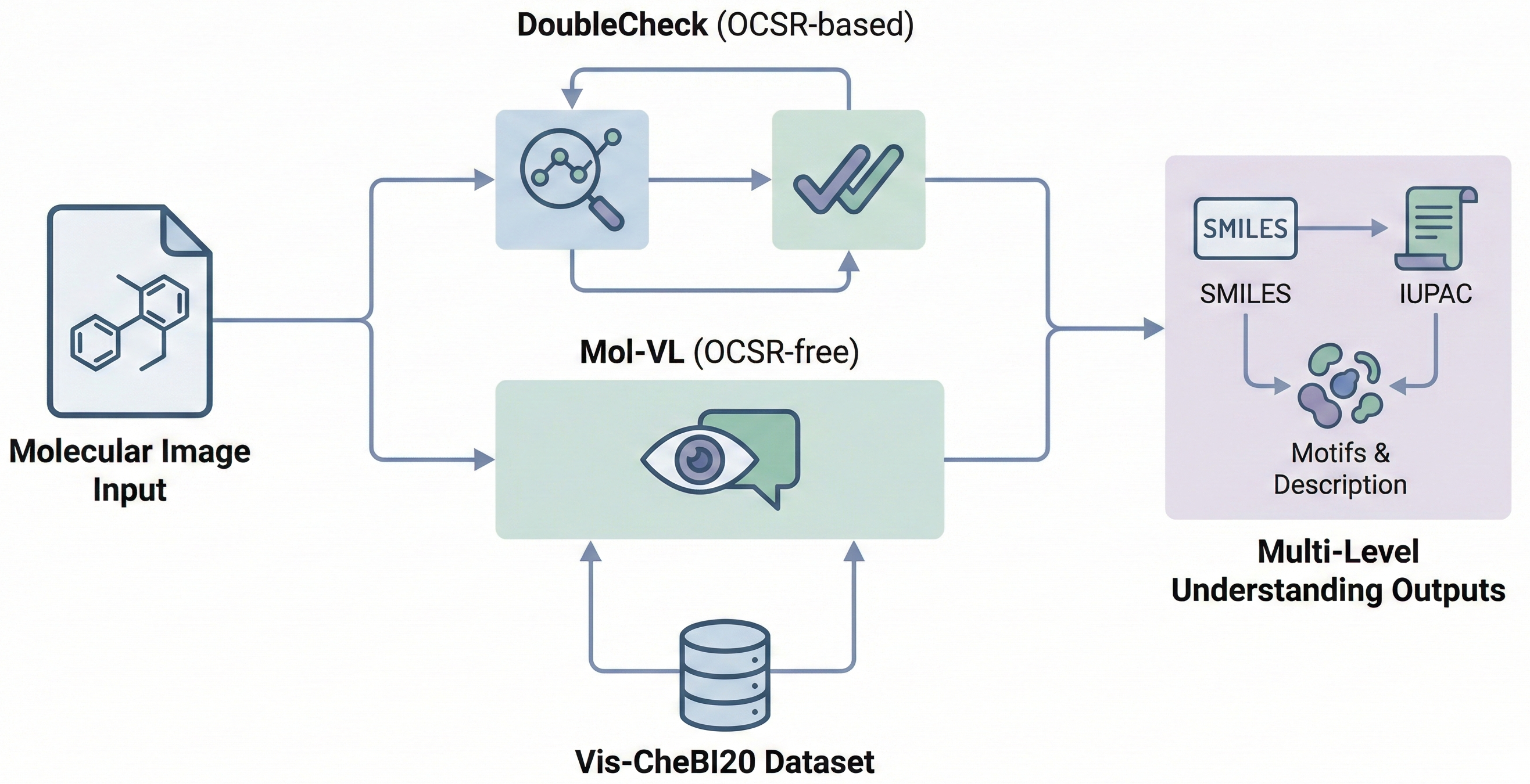
- Methodological: It proposes two novel architectures, DoubleCheck (an enhanced recognition model) and Mol-VL (an end-to-end vision-language model), to solve the newly formulated OCSU task.
- Resource: It constructs and releases Vis-CheBI20, the first large-scale dataset specifically designed for optical chemical structure understanding, containing 29.7K images and 117.7K image-text pairs.
What is the motivation?
Existing methods for processing molecular images focus narrowly on Optical Chemical Structure Recognition (OCSR), which translates an image solely into a machine-readable graph or SMILES string. However, SMILES strings are not chemist-friendly and lack high-level semantic context.
- Gap: There is a lack of systems that can translate chemical diagrams into human-readable descriptions (e.g., functional groups, IUPAC names) alongside the graph structure.
- Goal: To enable Optical Chemical Structure Understanding (OCSU), bridging the gap between visual representations and both machine/chemist-readable descriptions to support drug discovery and property prediction.
What is the novelty here?
The paper introduces the OCSU task, enabling multi-level understanding (motif, molecule, and abstract levels). To solve this, it introduces two distinct paradigms:
- DoubleCheck (OCSR-based): An enhancement to standard OCSR models (like MolScribe) that performs a “second look” at locally ambiguous atoms. It uses attentive feature enhancement to fuse global molecular features with local features from ambiguous regions.
- Mol-VL (OCSR-free): An end-to-end Vision-Language Model (VLM) based on Qwen2-VL. It uses multi-task learning to directly generate text descriptions from molecular images without an intermediate SMILES step.
- Vis-CheBI20 Dataset: A new benchmark specifically constructed for OCSU, deriving captions and functional group data from ChEBI-20 and PubChem.
What experiments were performed?
The authors evaluated both paradigms on Vis-CheBI20 and existing benchmarks (USPTO, ACS) across four subtasks:
- Functional Group Caption: Retrieval/F1 score evaluation.
- Molecule Description: Natural language generation metrics (BLEU, ROUGE, METEOR).
- IUPAC Naming: Text generation metrics (BLEU, ROUGE).
- SMILES Naming (OCSR): Exact matching accuracy ($Acc_s$).
Baselines:
- Task-Specific: MolScribe, MolVec, OSRA.
- LLM/VLM: Qwen2-VL, BioT5+, Mol-Instructions.
- Ablation: DoubleCheck vs. MolScribe backbone to test the “feature enhancement” mechanism.
What were the outcomes and conclusions drawn?
- DoubleCheck Superiority: DoubleCheck outperformed the MolScribe baseline on OCSR tasks, achieving 92.85% accuracy on Vis-CheBI20 (vs 92.57%) and showing significant gains on chiral molecules in the ACS dataset (+3.12%).
- Paradigm Trade-offs:
- Mol-VL (OCSR-free) excelled at semantic tasks like Functional Group Captioning, outperforming the strongest baseline by 7.7% in F1 score. It benefits from end-to-end learning of structural context.
- DoubleCheck (OCSR-based) performed better on IUPAC naming recall and exact SMILES recovery, as explicit graph reconstruction is more precise for rigid nomenclature than VLM generation.
- Conclusion: Enhancing submodules improves OCSR-based paradigms, while end-to-end VLMs offer stronger semantic understanding but struggle with exact syntax generation (SMILES/IUPAC).
Reproducibility Details
Data
Vis-CheBI20 Dataset
- Source: Derived from ChEBI-20 and PubChem.
- Size: 29,700 molecular diagrams, 117,700 image-text pairs.
- Generation: Images generated from SMILES using RDKit to simulate real-world journal/patent styles.
- Splits:
- Training: ~26,000 images (varies slightly by task).
- Test: ~3,300 images.
| Task | Train Size | Test Size |
|---|---|---|
| Functional Group | 26,144 | 3,269 |
| Description | 26,407 | 3,300 |
| IUPAC/SMILES | 26,200 | 2,680 |
Algorithms
DoubleCheck (Attentive Feature Enhancement)
- Ambiguity Detection: Uses atom prediction confidence to identify “ambiguous atoms”.
- Masking: Applies a 2D Gaussian mask to the image centered on the ambiguous atom.
- Local Encoding: A Swin-B encoder ($\Phi_l$) encodes the masked image region.
- Fusion: Aligns local features ($\mathcal{F}_l$) with global features ($\mathcal{F}_g$) using a 2-layer MLP and fuses them via weighted summation.
- Equation: $\mathcal{F}_e = \mathcal{F}_g + MLP(\mathcal{F}_g \oplus \hat{\mathcal{F}}_l) * \hat{\mathcal{F}}_l$.
- Two-Stage Training:
- Stage 1: Train atom/bond predictors (30 epochs).
- Stage 2: Train alignment/fusion modules with random Gaussian mask noise (10 epochs).
Mol-VL (Multi-Task VLM)
- Prompting: System prompt: “You are working as an excellent assistant in chemistry…”
- Tokens: Uses
<image>and</image>special tokens. - Auxiliary Task: Functional group recognition (identifying highlighted groups) added to training to improve context learning.
Models
- DoubleCheck:
- Backbone: MolScribe architecture.
- Encoders: Swin-B for both global and local atom encoding.
- Mol-VL:
- Base Model: Qwen2-VL (2B and 7B versions).
- Vision Encoder: ViT with naive dynamic resolution and M-ROPE.
Evaluation
Key Metrics:
- SMILES: Exact Match Accuracy ($Acc_s$), Chiral Accuracy ($Acc_c$).
- Functional Groups: F1 Score (Information Retrieval task).
- Text Generation: BLEU-2/4, METEOR, ROUGE-L.
Selected Results: | Model | Task | Metric | Score | | :— | :— | :— | :— | | DoubleCheck | OCSR (Vis-CheBI20) | $Acc_s$ | 92.85% | | MolScribe | OCSR (Vis-CheBI20) | $Acc_s$ | 92.57% | | Mol-VL-7B | Func. Group Caption | F1 | 97.32% | | DoubleCheck | Func. Group Caption | F1 | 93.63% |
Hardware
- DoubleCheck: Trained on 4 NVIDIA A100 GPUs for 4 days.
- Max LR: 4e-4.
- Mol-VL: Trained on 4 NVIDIA A100 GPUs for 10 days.
- Max LR: 1e-5, 50 epochs.
Citation
@misc{fanOCSUOpticalChemical2025,
title = {OCSU: Optical Chemical Structure Understanding for Molecule-centric Scientific Discovery},
shorttitle = {OCSU},
author = {Fan, Siqi and Xie, Yuguang and Cai, Bowen and Xie, Ailin and Liu, Gaochao and Qiao, Mu and Xing, Jie and Nie, Zaiqing},
year = {2025},
month = jan,
number = {arXiv:2501.15415},
eprint = {2501.15415},
primaryclass = {cs},
publisher = {arXiv},
doi = {10.48550/arXiv.2501.15415},
archiveprefix = {arXiv}
}