Paper Information
Citation: Zhang, W., Wang, X., Feng, B., & Liu, W. (2025). MolSight: Optical Chemical Structure Recognition with SMILES Pretraining, Multi-Granularity Learning and Reinforcement Learning. arXiv preprint arXiv:2511.17300. https://doi.org/10.48550/arXiv.2511.17300
Publication: arXiv 2025
Additional Resources:
What kind of paper is this?
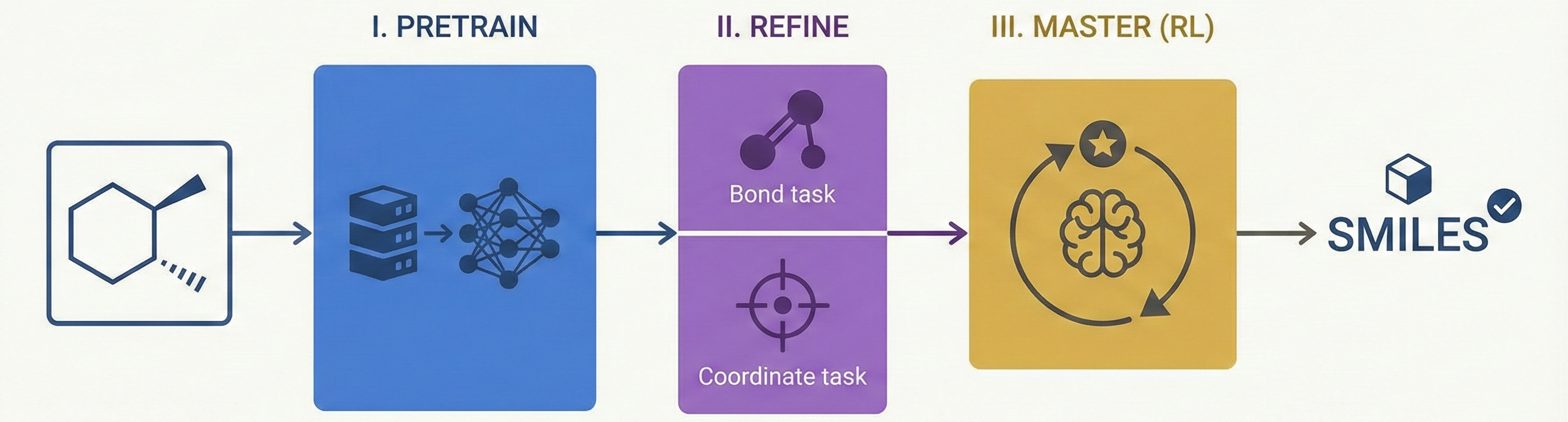
This is primarily a Method paper. It proposes a novel three-stage training framework (Pretraining → Fine-tuning → RL Post-training) to improve Optical Chemical Structure Recognition (OCSR). Specifically, it introduces the use of Group Relative Policy Optimization (GRPO) to solve non-differentiable chemical validity issues.
It also has a Resource component, as the authors construct and release Stereo-200k, a dataset specifically designed to train models on challenging stereoisomeric molecules.
What is the motivation?
Existing OCSR systems struggle to accurately recognize stereochemical information (e.g., chirality, geometric isomerism) because the visual cues distinguishing stereoisomers - such as wedge and dash bonds - are subtle. Current methods often fail to capture the geometric relationships required to distinguish molecules with identical connectivity but different spatial arrangements. Accurate recognition is critical for downstream tasks like drug discovery where stereochemistry determines pharmacological effects.
What is the novelty here?
MolSight introduces three key technical innovations:
- Reinforcement Learning for OCSR: It is the first OCSR system to incorporate RL (specifically GRPO) to directly optimize for chemical semantic correctness.
- Multi-Granularity Learning: It employs auxiliary heads for chemical bond classification and atom localization. Unlike previous approaches that optimize these jointly, MolSight decouples the coordinate head to prevent interference with SMILES generation.
- SMILES-M Notation: A lightweight extension to SMILES to handle Markush structures (common in patents) without significant sequence length increase.
What experiments were performed?
The authors evaluated MolSight using a rigorous mix of real and synthetic benchmarks:
- Baselines: Compared against rule-based (OSRA, MolVec) and deep learning methods (MolScribe, MolGrapher, DECIMER).
- Benchmarks: Evaluated on real-world datasets (USPTO, Maybridge UoB, CLEF-2012, JPO) and synthetic datasets (Staker, ChemDraw, Indigo, Stereo-2K).
- Ablation Studies: Tested the impact of the bond head, coordinate head, and RL stages separately.
- Transfer Learning: Assessed the quality of learned representations by using the frozen encoder for molecular property prediction on MoleculeNet.
What outcomes/conclusions?
- SOTA Performance: MolSight achieved 85.1% stereochemical accuracy on the USPTO dataset, significantly outperforming the previous SOTA (MolScribe) which achieved 69.0%.
- RL Effectiveness: Reinforcement learning post-training specifically improved performance on stereoisomers, raising Tanimoto similarity and exact match rates on the Stereo-2k test set.
- Robustness: The model maintained high performance even on perturbed (rotated/sheared) and low-resolution images, outperforming rule-based methods significantly in these scenarios.
Reproducibility Details
Data
The training pipeline uses three distinct data sources:
- Pre-training: MolParser-7M. Contains diverse images but requires the SMILES-M extension to handle Markush structures.
- Fine-tuning: PubChem-1M and USPTO-680K. Used for multi-granularity learning with bond and coordinate labels.
- RL Post-training: Stereo-200k. A self-collected dataset from the first 2M compounds in PubChem, filtered for chirality (’@’) and cis-trans isomerism (’/’, ‘\’). It uses 5 different RDKit drawing styles to ensure robustness.
Algorithms
- Reinforcement Learning: Uses GRPO (Group Relative Policy Optimization).
- Reward Function: A linear combination of Tanimoto similarity ($w_t=0.4$) and Stereochemical exact match ($w_s=0.6$).
- Sampling: Samples 4 completions per image with temperature 1.0 during RL training.
- Auxiliary Tasks:
- Bond Classification: Concatenates hidden states of two atom queries to predict bond type via MLP.
- Atom Localization: Treated as a classification task (SimCC) but optimized using Maximum Likelihood Estimation (MLE) to account for uncertainty.
Models
- Architecture: Encoder-Decoder Transformer.
- Encoder: EfficientViT-L1 (~53M params), chosen for linear attention efficiency.
- Decoder: 6-layer Transformer with RoPE, SwiGLU, and RMSNorm. Randomly initialized (no LLM weights) due to vocabulary mismatch.
- Coordinate Head: Separated from the main decoder. It adds 2 extra Transformer layers to process atom queries before prediction to improve accuracy.
- Parameter Tuning:
- Stage 3 (RL) uses LoRA (Rank=8, Alpha=16) to optimize the decoder.
Evaluation
- Metrics:
- Exact Match: String identity for standard SMILES.
- Tanimoto Coefficient: Fingerprint similarity for chemical semantics.
- OKS (Object Keypoint Similarity): Used specifically for evaluating atom localization accuracy.
- Perturbation: Robustness tested with random rotations [-5°, 5°] and xy-shearing [-0.1, 0.1].
Hardware
- Compute: Training and inference performed on a single node.
- Processors: Intel Xeon Silver 4210R CPU.
- Accelerators: 4x NVIDIA GeForce RTX 3090/4090 GPUs.
- Hyperparameters:
- Stage 1: Batch size 512, LR $4 \x10^{-4}$.
- Stage 2: Batch size 256, Bond head LR $4 \x10^{-4}$, Coord head LR $4 \x10^{-5}$.
- Stage 3 (RL): Batch size 64, Base LR $1 \x10^{-4}$.
Citation
@misc{zhang2025molsight,
title={MolSight: Optical Chemical Structure Recognition with SMILES Pretraining, Multi-Granularity Learning and Reinforcement Learning},
author={Wenrui Zhang and Xinggang Wang and Bin Feng and Wenyu Liu},
year={2025},
eprint={2511.17300},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2511.17300},
}