Key Contribution
Introduces MolParser-7M, the largest Optical Chemical Structure Recognition (OCSR) dataset, uniquely combining diverse synthetic data with a large volume of manually-annotated, “in-the-wild” images from real scientific documents to improve model robustness. Also introduces WildMol, a new challenging benchmark for evaluating OCSR performance on real-world data, including Markush structures.
Overview
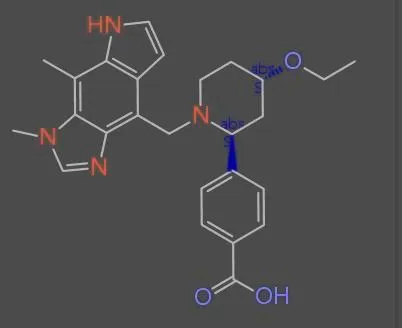
The MolParser project addresses the challenge of recognizing molecular structures from images found in real-world scientific documents. Unlike existing OCSR datasets that rely primarily on synthetically generated images, MolParser-7M incorporates 400,000 manually annotated images cropped from actual patents and scientific papers, making it the first large-scale dataset to bridge the gap between synthetic training data and real-world deployment scenarios.
Strengths
- Largest open-source OCSR dataset with over 7.7 million pairs
- The only large-scale OCSR training set that includes a significant amount (400k) of “in-the-wild” data cropped from real patents and literature
- High diversity of molecular structures from numerous sources (PubChem, ChEMBL, polymers, etc.)
- Introduces the WildMol benchmark for evaluating performance on challenging, real-world data, including Markush structures
- The “in-the-wild” fine-tuning data (MolParser-SFT-400k) was curated via an efficient active learning data engine with human-in-the-loop validation
Limitations
- The E-SMILES format cannot represent certain complex cases, such as coordination bonds, dashed abstract rings, and Markush structures depicted with special patterns
- The model and data do not yet fully exploit molecular chirality, which is critical for chemical properties
- Performance could be further improved by scaling up the amount of real annotated training data
Technical Notes
Synthetic Data Generation
To ensure diversity, molecular structures were collected from databases like ChEMBL, PubChem, and Kaggle BMS. A significant number of Markush, polymer, and fused-ring structures were also randomly generated. Images were rendered using RDKit and epam.indigo with randomized parameters (e.g., bond width, font size, rotation) to increase visual diversity.
In-the-Wild Data Engine (MolParser-SFT-400k)
A YOLOv11 object detection model (MolDet) located and cropped over 20 million molecule images from 1.22 million real PDFs (patents and papers). After de-duplication via p-hash similarity, 4 million unique images remained.
An active learning algorithm was used to select the most informative samples for annotation, targeting images where an ensemble of models showed moderate confidence (0.6-0.9 Tanimoto similarity), indicating they were challenging but learnable.
This active learning approach with model pre-annotations reduced manual annotation time per molecule to 30 seconds, a 90% savings compared to annotating from scratch.