Paper Information
Citation: Chun, S., Kim, J., Jo, A., Jo, Y., & Oh, S. (2025). MolMole: Molecule Mining from Scientific Literature. arXiv preprint arXiv:2505.03777. https://doi.org/10.48550/arXiv.2505.03777
Publication: arXiv 2025
Additional Resources:
What kind of paper is this?
This is primarily a Method paper, with a strong Resource contribution.
It functions as a Method paper because it introduces “MolMole,” a unified deep learning framework that integrates molecule detection, reaction diagram parsing, and optical chemical structure recognition (OCSR) into a single pipeline. It validates this method through extensive comparisons against state-of-the-art baselines like DECIMER and OpenChemIE.
It also serves as a Resource paper because the authors construct and release a novel page-level benchmark dataset of 550 annotated pages (patents and articles) to address the lack of standardized evaluation metrics for full-page chemical extraction.
What is the motivation?
The rapid accumulation of chemical literature has trapped valuable molecular and reaction data in unstructured formats like images and PDFs. Extracting this manually is time-consuming, while existing AI frameworks have significant limitations:
- DECIMER: Lacks the ability to process reaction diagrams entirely.
- OpenChemIE: Relies on external layout parser models to crop elements before processing. This dependence often leads to detection failures in documents with complex layouts.
- Generative Hallucination: Existing generative OCSR models (like MolScribe) are prone to “hallucinating” structures or failing on complex notations like polymers.
What is the novelty here?
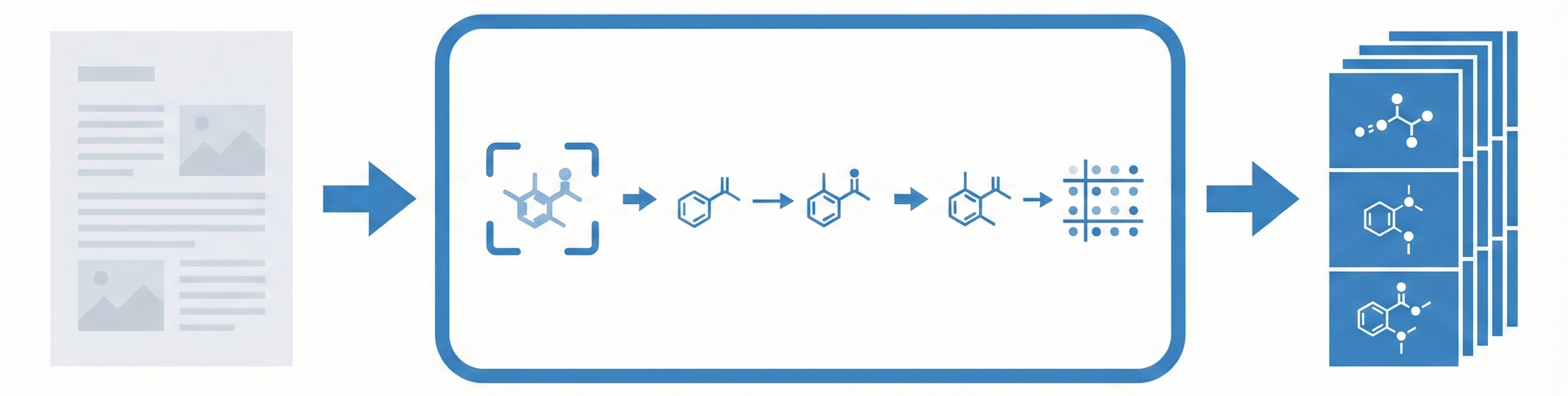
MolMole introduces several architectural and workflow innovations:
- Direct Page-Level Processing: Unlike OpenChemIE, MolMole processes full document pages directly without requiring an external layout parser, which improves robustness on complex layouts like two-column patents.
- Unified Vision Pipeline: It integrates three specialized vision models into one workflow:
- ViDetect: A DINO-based object detector for identifying molecular regions.
- ViReact: An RxnScribe-based model adapted for full-page reaction parsing.
- ViMore: A detection-based OCSR model that explicitly predicts atoms and bonds.
- Hallucination Mitigation: By using a detection-based approach (ViMore), the model avoids hallucinating chemical structures and provides confidence scores.
- Advanced Notation Support: The system explicitly handles “wavy bonds” (variable attachments in patents) and polymer bracket notations, which confuse standard SMILES-based models.
What experiments were performed?
The authors evaluated the framework on both a newly curated benchmark and existing public datasets:
- New Benchmark Creation: They curated 550 pages (300 patents, 250 articles) fully annotated with bounding boxes, reaction roles (reactant, product, condition), and MOLfiles.
- Baselines: MolMole was compared against DECIMER 2.0, OpenChemIE, and ReactionDataExtractor 2.0.
- OCSR Benchmarking: ViMore was evaluated against DECIMER, MolScribe, and MolGrapher on four public datasets: USPTO, UOB, CLEF, and JPO.
- Metric Proposal: They introduced a combined “End-to-End” metric that modifies standard object detection Precision/Recall to strictly require correct SMILES conversion for a “True Positive”.
What outcomes/conclusions?
- SOTA Page-Level Performance: On the new benchmark, MolMole achieved F1 scores of 89.1% (Patents) and 86.8% (Articles) for the combined detection-to-conversion task, significantly outperforming DECIMER and OpenChemIE.
- Reaction Parsing: ViReact achieved an F1 score of 98.0% (soft match) on patents, compared to 82.2% for the next best model (RxnScribe).
- Public Benchmarks: ViMore outperformed competitors on 3 out of 4 public OCSR datasets (CLEF, JPO, USPTO).
- Qualitative Superiority: The authors demonstrated that MolMole successfully handles multi-column reaction diagrams where cropping-based models fail and faithfully preserves layout geometry in generated MOLfiles.
Reproducibility Details
Data
- Training Data: The models (ViDetect and ViMore) were trained on private/proprietary datasets, which is a limitation for full reproducibility from scratch.
- Benchmark Data: The authors introduce a test set of 550 pages (3,897 molecules, 1,022 reactions) derived from patents and scientific articles. This dataset is stated to be made “publicly available”.
- Public Evaluation Data: Standard OCSR datasets used include USPTO (5,719 images), UOB (5,740 images), CLEF (992 images), and JPO (450 images).
Algorithms
- Pipeline Workflow: PDF → PNG Images → Parallel execution of ViDetect and ViReact → Cropping of molecular regions → ViMore conversion → Output (JSON/Excel).
- Post-Processing:
- ViDetect: Removes overlapping proposals based on confidence scores and size constraints.
- ViReact: Refines predictions by correcting duplicates and removing empty entities.
- ViMore: Assembles detected atom/bond information into structured representations (MOLfile).
Models
| Model | Architecture Basis | Task | Key Feature |
|---|---|---|---|
| ViDetect | DINO (DETR-based) | Molecule Detection | End-to-end training; avoids slow autoregressive methods. |
| ViReact | RxnScribe | Reaction Parsing | Operates on full pages; autoregressive decoder for structured sequence generation. |
| ViMore | Custom Vision Model | OCSR | Detection-based (predicts atom/bond regions). |
Evaluation
- Molecule Detection: Evaluated using COCO metrics (AP, AR, F1) at IoU thresholds 0.50-0.95.
- Molecule Conversion: Evaluated using SMILES exact match accuracy and Tanimoto similarity.
- Combined Metric: A custom metric where a True Positive requires both IoU ≥ 0.5 and a correct SMILES string match ($SMILES_{gt} == SMILES_{pred}$).
- Reaction Parsing: Evaluated using Hard Match (all components correct) and Soft Match (molecular entities only, ignoring text labels).