Paper Information
Citation: Zhang, D., Zhao, D., Wang, Z., Li, J., & Li, J. (2024). MMSSC-Net: multi-stage sequence cognitive networks for drug molecule recognition. RSC Advances, 14(26), 18182-18191. https://doi.org/10.1039/D4RA02442G
Publication: RSC Advances 2024
What kind of paper is this?
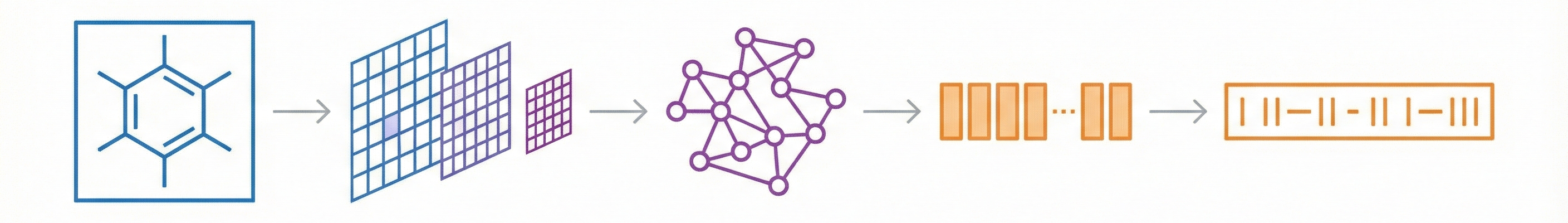
Methodological Paper ($\Psi_{\text{Method}}$). The paper proposes a novel deep learning architecture (MMSSC-Net) for Optical Chemical Structure Recognition (OCSR). It focuses on architectural innovation - combining a SwinV2 visual encoder with a GPT-2 decoder - and validates this method through extensive benchmarking against existing rule-based and deep-learning baselines. It includes ablation studies to justify the choice of the visual encoder.
What is the motivation?
- Data Usage Gap: Drug discovery relies heavily on scientific literature, but molecular structures are often locked in vector graphics or images that computers cannot easily process.
- Limitations of Prior Work: Existing Rule-based methods are rigid and sensitive to noise. Previous Deep Learning approaches (Encoder-Decoder “Image Captioning” styles) often lack precision, interpretability, and struggle with varying image resolutions or large molecules.
- Need for “Cognition”: The authors argue that treating the image as a single isolated whole is insufficient; a model needs to “perceive” fine-grained details (atoms and bonds) to handle noise and varying pixel qualities effectively.
What is the novelty here?
- Multi-Stage Cognitive Architecture: MMSSC-Net splits the task into stages:
- Fine-grained Perception: Detecting atom and bond sequences (including spatial coordinates) using SwinV2.
- Graph Construction: Assembling these into a molecular graph.
- Sequence Evolution: converting the graph into a machine-readable format (SMILES).
- Hybrid Transformer Model: It combines a hierarchical vision transformer (SwinV2) for encoding with a generative pre-trained transformer (GPT-2) and MLPs for decoding atomic and bond targets.
- Robustness Mechanisms: The inclusion of random noise sequences during training to improve generalization to new molecular targets.
What experiments were performed?
- Baselines: compared against 7 other tools:
- Rule-based: MolVec, OSRA.
- Image-Smiles (DL): ABC-Net, Img2Mol, MolMiner.
- Image-Graph-Smiles (DL): Image-To-Graph, MolScribe, ChemGrapher.
- Datasets: Evaluated on 5 diverse datasets: STAKER (synthetic), USPTO, CLEF, JPO, and UOB (real-world).
- Metrics:
- Accuracy: Exact string match of the predicted SMILES.
- Tanimoto Similarity: Chemical similarity using Morgan fingerprints.
- Ablation Study: Tested different visual encoders (Swin Transformer, ViT-B, ResNet-50) to validate the choice of SwinV2.
- Resolution Sensitivity: Tested model performance across image resolutions from 256px to 2048px.
What were the outcomes and conclusions drawn?
- State-of-the-Art Performance: MMSSC-Net achieved 75-94% accuracy across datasets, outperforming baselines on most benchmarks.
- Resolution Robustness: The model maintained high accuracy (approx 90-95%) across resolutions (256px to 2048px), whereas baselines like Img2Mol dropped significantly at higher resolutions.
- Efficiency: The SwinV2 encoder was noted to be more efficient than ViT-B in this context.
- Limitations: The model struggles with stereochemistry (virtual vs. solid wedge bonds) and “irrelevant text” noise (e.g., in JPO/DECIMER datasets).
Reproducibility Details
Data
The model was trained on a combination of PubChem and USPTO data, augmented to handle visual variability.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Training | PubChem | 1,000,000 | Converted from InChI to SMILES; random sampling. |
| Training | USPTO | 600,000 | Patent images; converted from MOL to SMILES. |
| Evaluation | STAKER | 40,000 | Synthetic; Avg res $256 \x256$. |
| Evaluation | USPTO | 4,862 | Real; Avg res $721 \x432$. |
| Evaluation | CLEF | 881 | Real; Avg res $1245 \x412$. |
| Evaluation | JPO | 380 | Real; Avg res $614 \x367$. |
| Evaluation | UOB | 5,720 | Real; Avg res $759 \x416$. |
Augmentation:
- Image: Random perturbations using RDKit/Indigo (rotation, filling, cropping, bond thickness/length, font size, Gaussian noise).
- Molecular: Introduction of functional group abbreviations and R-substituents (dummy atoms) using SMARTS templates.
Algorithms
- Target Sequence Formulation: The model predicts a sequence containing bounding box coordinates and type labels: $\{y_{min}, x_{min}, y_{max}, x_{max}, C_{type}\}$.
- Loss Function: Cross-entropy loss with maximum likelihood estimation. $$\max \sum_{i=1}^{N} \sum_{j=1}^{L} \omega_{j} \log P(t_{j}^{i}|x^{i}, t_{1}^{i}, \dots, t_{j-1}^{i})$$
- Noise Injection: A random sequence $T_r$ is appended to the target sequence during training to improve generalization to new goals.
- Graph Construction: Atoms ($v$) and bonds ($e$) are recognized separately; bonds are defined by connecting spatial atomic coordinates.
Models
- Encoder: Swin Transformer V2.
- Pre-trained on ImageNet-1K.
- Window size: $16 \x16$.
- Parameters: 88M.
- Input resolution: $256 \x256$.
- Features: Scaled cosine attention; log-space continuous position bias.
- Decoder: GPT-2 + MLP.
- GPT-2: Used for recognizing atom types.
- Layers: 24.
- Attention Heads: 12.
- Hidden Dimension: 768.
- Dropout: 0.1.
- MLP: Used for classifying bond types (single, double, triple, aromatic, wedge).
- GPT-2: Used for recognizing atom types.
- Vocabulary:
- Standard: 95 common numbers/characters ([0], [C], [=], etc.).
- Extended: 2000 SMARTS-based characters for isomers/groups (e.g., “[C2F5]”, “[halo]”).
Evaluation
Metrics:
- Accuracy: Exact match of the generated SMILES string.
- Tanimoto Similarity: Similarity of Morgan fingerprints between predicted and ground truth molecules.
Key Results (Accuracy):
| Dataset | MMSSC-Net | MolVec (Rule) | ABC-Net (DL) | MolScribe (DL) |
|---|---|---|---|---|
| Indigo | 98.14 | 95.63 | 96.4 | 99.0 |
| USPTO | 94.24 | 88.47 | * | 51.7 |
| CLEF | 91.26 | 81.61 | 96.1 | 82.9 |
| UOB | 92.71 | 81.32 | * | 86.9 |
Hardware
- Training Configuration:
- Batch Size: 128.
- Learning Rate: $4 \x10^{-5}$.
- Epochs: 40.
- Inference Speed: The SwinV2 encoder demonstrated higher efficiency (faster inference time) compared to ViT-B and ResNet-50 baselines during ablation.
Citation
@article{zhangMMSSCNetMultistageSequence2024,
title = {MMSSC-Net: Multi-Stage Sequence Cognitive Networks for Drug Molecule Recognition},
shorttitle = {MMSSC-Net},
author = {Zhang, Dehai and Zhao, Di and Wang, Zhengwu and Li, Junhui and Li, Jin},
year = 2024,
journal = {RSC Advances},
volume = {14},
number = {26},
pages = {18182--18191},
publisher = {Royal Society of Chemistry},
doi = {10.1039/D4RA02442G},
url = {https://pubs.rsc.org/en/content/articlelanding/2024/ra/d4ra02442g}
}