Paper Information
Citation: Morin, L., Weber, V., Nassar, A., Meijer, G. I., Van Gool, L., Li, Y., & Staar, P. (2025). MarkushGrapher: Joint Visual and Textual Recognition of Markush Structures. 2025 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 14505-14515. https://doi.org/10.1109/CVPR52734.2025.01352
Publication: CVPR 2025
Additional Resources:
What kind of paper is this?
This is primarily a Method paper, as it proposes a novel neural architecture (MarkushGrapher) and a specialized data representation (Optimized CXSMILES) to solve the problem of Markush structure recognition. It also has a strong Resource component, as it introduces a synthetic data generation pipeline and “M2S,” the first annotated benchmark of real-world Markush structures from patent documents.
What is the motivation?
The automated analysis of chemical literature, particularly patents, is critical for drug discovery and material science. A major bottleneck is the extraction of Markush structures - complex chemical templates that represent families of molecules using a core backbone image and textual variable definitions. Existing methods are limited because they either focus solely on images (OCSR), missing the textual context, or solely on text, missing the structural backbone. There is a practical need for a unified, multi-modal approach that can jointly interpret visual and textual data to accurately extract these structures for prior-art search and database construction.
What is the novelty here?
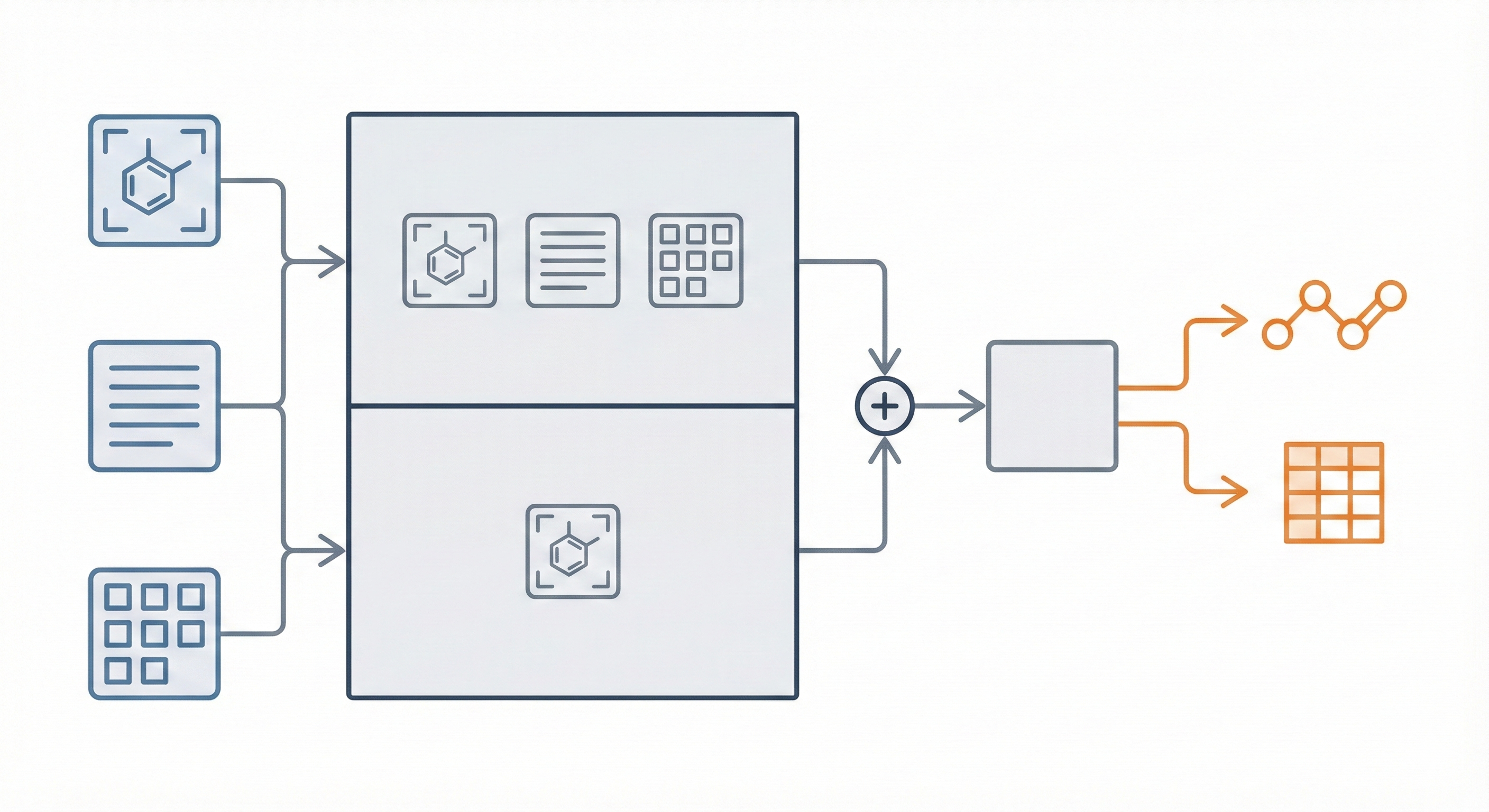
The core novelty is MarkushGrapher, a multi-modal architecture that jointly encodes image, text, and layout information. Key contributions include:
- Dual-Encoder Architecture: Combines a Vision-Text-Layout (VTL) encoder (based on UDOP) with a specialized, pre-trained Optical Chemical Structure Recognition (OCSR) encoder (MolScribe).
- Joint Recognition: The model autoregressively generates a sequential graph representation (Optimized CXSMILES) and a substituent table simultaneously, allowing it to leverage cross-modal dependencies (e.g., using text to infer bond types in the image).
- Synthetic Data Pipeline: A comprehensive pipeline to generate realistic synthetic Markush structures (images and text) from PubChem data, overcoming the lack of labeled training data.
- Optimized Representation: A compacted version of CXSMILES that moves variable groups into the SMILES string and adds explicit atom indexing to handle complex “frequency” and “position” variation indicators.
What experiments were performed?
The authors validated their approach using the following setup:
- Baselines: Compared against image-only chemistry models (DECIMER, MolScribe) and general-purpose multi-modal models (Uni-SMART, GPT-4o, Pixtral, Llama-3.2).
- Datasets: Evaluated on three benchmarks:
- MarkushGrapher-Synthetic: 1,000 generated samples.
- M2S: A new benchmark of 103 manually annotated real-world patent images.
- USPTO-Markush: 74 Markush backbone images from USPTO patents.
- Ablation Studies: Analyzed the impact of the OCSR encoder, fusion strategies (early vs. late), and the optimized CXSMILES format.
What outcomes/conclusions?
- SOTA Performance: MarkushGrapher significantly outperformed all baselines. On the M2S benchmark, it achieved a 38% Exact Match on CXSMILES (vs. 21% for MolScribe) and 29% Exact Match on tables (vs. 0-19% for others).
- Feature Handling: It is the only method capable of reliably recognizing complex Markush features like frequency variation (‘Sg’) and position variation (’m’) indicators, where other models scored near zero.
- Cross-Modal Reasoning: Qualitative analysis showed the model can correctly infer visual details (like bond order) that are ambiguous in the image but clarified by the text description.
- Robustness: The model generalized well to real-world data despite being trained on synthetic data.
Reproducibility Details
Data
Training Data
- Source: Synthetic dataset generated from PubChem SMILES.
- Size: 210,000 synthetic images.
- Pipeline:
- Selection: Sampled SMILES from PubChem based on substructure diversity.
- Augmentation: SMILES augmented to artificial CXSMILES using RDKit (inserting variable groups, frequency indicators).
- Rendering: Images rendered using Chemistry Development Kit (CDK) with randomized drawing parameters (font, bond width, spacing).
- Text Generation: Textual definitions generated using manual templates extracted from patents; 10% were paraphrased using Mistral-7B-Instruct-v0.3 to increase diversity.
- OCR: Bounding boxes extracted via a custom SVG parser aligned with MOL files.
Evaluation Data
- M2S Dataset: 103 images from USPTO, EPO, and WIPO patents (1999-2023), manually annotated with CXSMILES and substituent tables.
- USPTO-Markush: 74 images from USPTO patents (2010-2016).
- MarkushGrapher-Synthetic: 1,000 samples generated via the pipeline.
Algorithms
- Optimized CXSMILES:
- Compression: Variable groups moved from the extension block to the main SMILES string as special atoms to reduce sequence length.
- Indexing: Atom indices appended to each atom (e.g.,
C:1) to explicitly link the graph to the extension block (crucial formandSgsections). - Vocabulary: Specific tokens used for atoms and bonds.
- Augmentation: Standard image augmentations (shift, scale, blur, pepper noise, random lines) and OCR text augmentations (character substitution/insertion/deletion).
Models
- Architecture: Encoder-Decoder Transformer.
- VTL Encoder: T5-large encoder (initialized from UDOP) that processes image patches, text tokens, and layout (bounding boxes).
- OCSR Encoder: Vision encoder from MolScribe (Swin Transformer), frozen during training.
- Text Decoder: T5-large decoder.
- Fusion Strategy: Late Fusion. The output of the VTL encoder ($e_1$) is concatenated with the projected output of the OCSR encoder ($e_2$) before being passed to the decoder.
- Parameters: 831M total (744M trainable).
Evaluation
Metrics:
- CXSMILES Exact Match (EM): Requires perfect match of SMILES string, variable groups,
msections, andSgsections (ignoring stereochemistry). - Tanimoto Score: Similarity of RDKit DayLight fingerprints (Markush features removed).
- Table Exact Match: All variable groups and substituents must match.
- Table F1-Score: Aggregated recall and precision of substituents per variable group.
Hardware
- Compute: Trained on a single NVIDIA H100 GPU.
- Training Config: 10 epochs, batch size of 10, learning rate 5e-4, 100 warmup steps, weight decay 1e-3.
Citation
@inproceedings{morinMarkushGrapherJointVisual2025,
title = {MarkushGrapher: Joint Visual and Textual Recognition of Markush Structures},
shorttitle = {MarkushGrapher},
booktitle = {2025 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)},
author = {Morin, Lucas and Weber, Valéry and Nassar, Ahmed and Meijer, Gerhard Ingmar and Van Gool, Luc and Li, Yawei and Staar, Peter},
year = {2025},
month = jun,
pages = {14505--14515},
doi = {10.1109/CVPR52734.2025.01352}
}