Paper Information
Citation: Li, D., Xu, X., Pan, J., Gao, W., & Zhang, S. (2024). Image2InChI: Automated Molecular Optical Image Recognition. Journal of Chemical Information and Modeling, 64(9), 3640-3649. https://doi.org/10.1021/acs.jcim.3c02082
Publication: Journal of Chemical Information and Modeling (JCIM) 2024
Additional Resources:
Note: These notes are based on the Abstract and Supporting Information files only.
What kind of paper is this?
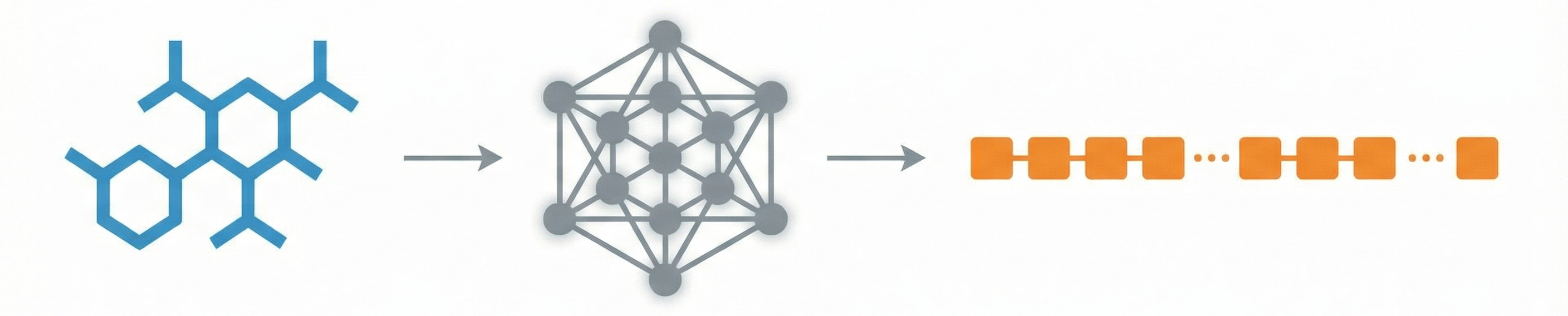
This is a Methodological Paper ($\Psi_{\text{Method}}$). It proposes a specific new deep learning architecture (“Image2InChI”) to solve the task of Optical Chemical Structure Recognition (OCSR). The rhetorical focus is on engineering a system that outperforms baselines on specific metrics (InChI accuracy, MCS accuracy) and providing a “valuable reference” for future algorithmic work.
What is the motivation?
The accurate digitization of chemical literature is a bottleneck in AI-driven drug discovery. Chemical structures in patents and papers exist as optical images (pixels), but machine learning models require machine-readable string representations (like InChI or SMILES). Efficiently and automatically bridging this gap is a prerequisite for large-scale data mining in chemistry.
What is the novelty here?
The core novelty is the Image2InChI architecture, which integrates:
- Improved SwinTransformer Encoder: Uses a hierarchical vision transformer to capture image features.
- Feature Fusion with Attention: A novel network designed to integrate image patch features with InChI prediction steps.
- End-to-End InChI Prediction: Unlike some methods that predict graph nodes/edges, this treats the problem as image-to-sequence translation targeting InChI strings directly.
What experiments were performed?
- Benchmark Validation: The model was trained and tested on the BMS1000 (Bristol-Myers Squibb) dataset from a Kaggle competition.
- Ablation/Comparative Analysis: The authors compared their method against other models in the supplement.
- Preprocessing Validation: They justified their choice of denoising algorithms (8-neighborhood vs. Gaussian/Mean) to ensure preservation of bond lines while removing “spiky point noise”.
What were the outcomes and conclusions drawn?
- High Accuracy: The model achieved 99.8% InChI accuracy, 94.8% Maximum Common Substructure (MCS) accuracy, and 96.2% Longest Common Subsequence (LCS) accuracy.
- Effective Denoising: The authors concluded that eight-neighborhood filtering is superior to mean or Gaussian filtering for this specific domain because it removes isolated noise points without blurring the fine edges of chemical bonds.
- Open Source: The authors committed to releasing the code to facilitate transparency and further research.
Reproducibility Details
Data
The primary dataset used is the BMS (Bristol-Myers Squibb) Dataset.
| Property | Details |
|---|---|
| Source | Kaggle Competition (BMS-Molecular-Translation) |
| Total Size | 2.4 million images |
| Training Set | 1.8 million images |
| Test Set | 0.6 million images |
| Content | Each image corresponds to a unique International Chemical Identifier (InChI) |
Other Datasets: The authors also utilized JPO (Japanese Patent Office), CLEF (CLEF-IP 2012), UOB (MolrecUOB), and USPTO datasets for broader benchmarking.
Preprocessing Pipeline:
- Denoising: Eight-neighborhood filtering (threshold < 4 non-white pixels) is used to remove salt-and-pepper noise while preserving bond lines. Mean and Gaussian filtering were rejected due to blurring.
- Sequence Padding:
- Analysis showed max InChI length < 270.
- Fixed sequence length set to 300.
- Tokens:
<sos>(190),<eos>(191),<pad>(192) used for padding/framing.
- Numerization: Characters are mapped to integers based on a fixed vocabulary (e.g., ‘C’ -> 178, ‘H’ -> 182).
Algorithms
Eight-Neighborhood Filtering (Denoising):
Pseudocode logic:
- Iterate through every pixel.
- Count non-white neighbors in the 3x3 grid (8 neighbors).
- If count < threshold (default 4), treat as noise and remove.
InChI Tokenization:
- InChI strings are split into character arrays.
- Example: Vitamin C
InChI=1S/C6H8O6...becomes[<sos>, C, 6, H, 8, O, 6, ..., <eos>, <pad>...]. - Mapped to integer tensor for model input.
Models
Architecture: Image2InChI
- Encoder: Improved SwinTransformer (Hierarchical Vision Transformer).
- Decoder: Transformer Decoder with patch embedding.
- Fusion: A novel “feature fusion network with attention” integrates the visual tokens with the sequence generation process.
- Framework: PyTorch 1.8.1.
Evaluation
Metrics:
- InChI Acc: Exact match accuracy of the predicted InChI string (Reported: 99.8%).
- MCS Acc: Maximum Common Substructure accuracy (structural similarity) (Reported: 94.8%).
- LCS Acc: Longest Common Subsequence accuracy (string similarity) (Reported: 96.2%).
- Morgan FP: Morgan Fingerprint similarity (Reported: 94.1%).
Hardware
| Component | Specification |
|---|---|
| GPU | NVIDIA Tesla P100 (16GB VRAM) |
| Platform | MatPool cloud platform |
| CPU | Intel Xeon Gold 6271 |
| RAM | 32GB System Memory |
| Driver | NVIDIA-SMI 440.100 |
| OS | Ubuntu 18.04 |
Citation
@article{li2024image2inchi,
title={Image2InChI: Automated Molecular Optical Image Recognition},
author={Li, Da-zhou and Xu, Xin and Pan, Jia-heng and Gao, Wei and Zhang, Shi-rui},
journal={Journal of Chemical Information and Modeling},
volume={64},
number={9},
pages={3640--3649},
year={2024},
publisher={American Chemical Society},
doi={10.1021/acs.jcim.3c02082}
}