Paper Information
Citation: Li, Y., Chen, G., & Li, X. (2022). Automated Recognition of Chemical Molecule Images Based on an Improved TNT Model. Applied Sciences, 12(2), 680. https://doi.org/10.3390/app12020680
Publication: MDPI Applied Sciences 2022
Additional Resources:
What kind of paper is this?
This is a Method paper.
It proposes a novel neural network architecture—the Image Captioning Model based on Deep TNT (ICMDT)—to solve the specific problem of “molecular translation” (image-to-text). The classification is supported by the following rhetorical indicators:
- Novel Mechanism: It introduces the “Deep TNT block” to improve upon the existing TNT architecture by fusing features at three levels (pixel, small patch, large patch).
- Baseline Comparison: The authors explicitly compare their model against four other architectures (CNN+RNN and CNN+Transformer variants).
- Ablation Study: Section 4.3 is dedicated to ablating specific components (position encoding, patch fusion) to prove their contribution to the performance gain.
What is the motivation?
The primary motivation is to speed up chemical research by digitizing historical chemical literature.
- Problem: Historical sources often contain corrupted or noisy images, making automated recognition difficult.
- Gap: Existing models like the standard TNT (Transformer in Transformer) function primarily as encoders for classification and fail to effectively integrate local pixel-level information required for precise structure generation.
- Goal: To build a dependable generative model that can accurately translate these noisy images into InChI (International Chemical Identifier) text strings.
What is the novelty here?
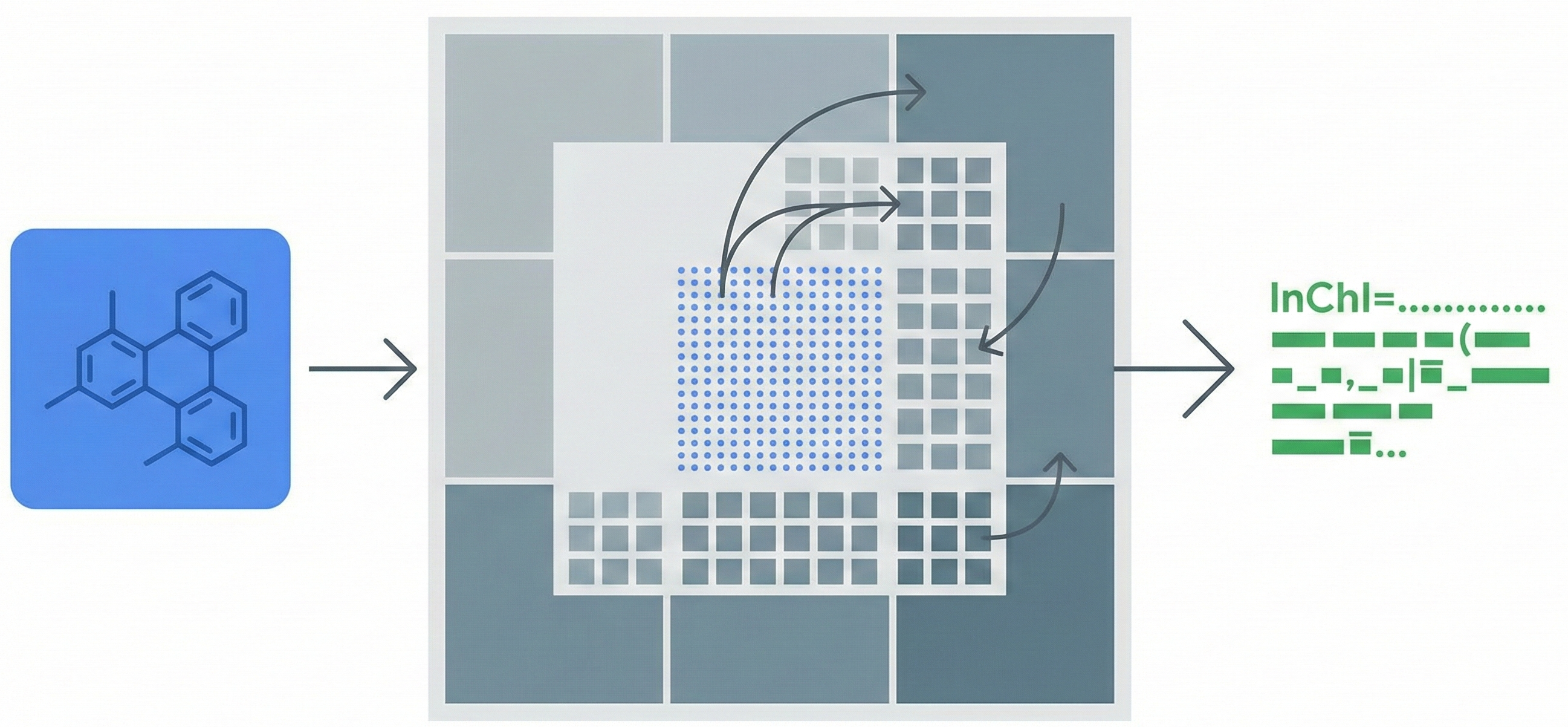
The core contribution is the Deep TNT block and the resulting ICMDT architecture.
- Deep TNT Block: Unlike the standard TNT which models local and global info, the Deep TNT block stacks three transformer blocks to process information at three granularities:
- Internal Transformer: Processes pixel embeddings.
- Middle Transformer: Processes small patch embeddings.
- Exterior Transformer: Processes large patch embeddings.
- Multi-level Fusion: The model fuses pixel-level features into small patches, and small patches into large patches, allowing for finer integration of local details.
- Position Encoding: A specific strategy of applying shared position encodings to small patches and pixels, while using a learnable 1D encoding for large patches.
What experiments were performed?
The authors evaluated the model on the Bristol-Myers Squibb-Molecular Translation dataset.
- Baselines: They constructed four comparative models:
- EfficientNetb0 + RNN (Bi-LSTM)
- ResNet50d + RNN (Bi-LSTM)
- EfficientNetb0 + Transformer
- ResNet101d + Transformer
- Ablation: They tested the impact of removing the large patch position encoding (ICMDT*) and reverting the encoder to a standard TNT-S (TNTD).
- Pre-processing Study: They experimented with denoising ratios and cropping strategies.
What were the outcomes and conclusions drawn?
- Performance: ICMDT achieved the lowest Levenshtein distance (0.69) compared to the best baseline (1.45 for ResNet101d+Transformer).
- Convergence: The model converged significantly faster than the baselines, outperforming others as early as epoch 6.7.
- Ablation Results: The full Deep TNT block reduced error by nearly half compared to the standard TNT encoder (0.69 vs 1.29 Levenshtein distance).
- Limitations: The model struggles with stereochemical layers (e.g., identifying clockwise neighbors or +/- signs) compared to non-stereochemical layers.
- Future Work: Integrating full object detection to predict atom/bond coordinates to better resolve 3D stereochemical information.
Reproducibility Details
Data
The experiments used the Bristol-Myers Squibb – Molecular Translation dataset from Kaggle.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Training | BMS Training Set | 2,424,186 images | Supervised; contains noise and blur |
| Evaluation | BMS Test Set | 1,616,107 images | Higher noise variation than training set |
Pre-processing Strategy:
- Effective: Padding resizing (reshaping to square, padding with border pixels).
- Ineffective: Smart cropping (removing white borders degraded performance).
- Augmentation: GaussNoise, Blur, RandomRotate90, and PepperNoise ($SNR=0.996$).
- Denoising: Best results found by mixing denoised and original data (Ratio 2:13) during training.
Algorithms
- Optimizer: Lookahead ($\alpha=0.5, k=5$) and RAdam ($\beta_1=0.9, \beta_2=0.99$).
- Loss Function: Anti-Focal loss ($\gamma=0.5$) combined with Label Smoothing.
- Training Schedule:
- Initial resolution: $224 \x224$.
- Fine-tuning: Resolution $384 \x384$ for labels $>150$ length.
- Batch size: Dynamic, increasing from 16 to 1024.
- Inference Strategy:
- Beam Search ($k=16$ initially, $k=64$ if failing InChI validation).
- Test Time Augmentation (TTA): Rotations of $90^\circ$.
- Ensemble: Step-wise logit ensemble and voting based on Levenshtein distance scores.
Models
ICMDT Architecture:
- Encoder (Deep TNT):
- Internal Block: Hidden size 160, Heads 4, MLP act GELU.
- Middle Block: Hidden size 640, Heads 10.
- Exterior Block: Hidden size 2560, Heads 16.
- Patch Sizes: Small patch $16 \x16$, Pixel patch $4 \x4$.
- Decoder (Vanilla Transformer):
- Dimensions: 5120 (Hidden), 1024 (FFN).
- Depth: 3 layers, Heads: 8.
- Vocab Size: 193 (InChI tokens).
Evaluation
Metric: Levenshtein Distance (measures single-character edit operations between generated and ground truth InChI strings).
| Model | Levenshtein Distance | Params (M) | Convergence (Epochs) |
|---|---|---|---|
| ICMDT (Ours) | 0.69 | 138.16 | ~9.76 |
| TNTD (Ablation) | 1.29 | 114.36 | - |
| ResNet101d + Transformer | 1.45 | 302.02 | 14+ |
| ResNet50d + RNN | 3.86 | 90.6 | 14+ |
| EfficientNetb0 + RNN | 2.95 | 46.3 | 11+ |
Citation
@article{liAutomatedRecognitionChemical2022,
title = {Automated {{Recognition}} of {{Chemical Molecule Images Based}} on an {{Improved TNT Model}}},
author = {Li, Yanchi and Chen, Guanyu and Li, Xiang},
year = 2022,
month = jan,
journal = {Applied Sciences},
volume = {12},
number = {2},
pages = {680},
publisher = {Multidisciplinary Digital Publishing Institute},
issn = {2076-3417},
doi = {10.3390/app12020680}
}