Paper Information
Citation: Hu, J., Wu, H., Chen, M., Liu, C., Wu, J., Yin, S., Yin, B., Liu, C., Du, J., & Dai, L. (2023). Handwritten Chemical Structure Image to Structure-Specific Markup Using Random Conditional Guided Decoder. Proceedings of the 31st ACM International Conference on Multimedia (pp. 8114-8124). https://doi.org/10.1145/3581783.3612573
Publication: ACM Multimedia 2023
Additional Resources:
What kind of paper is this?
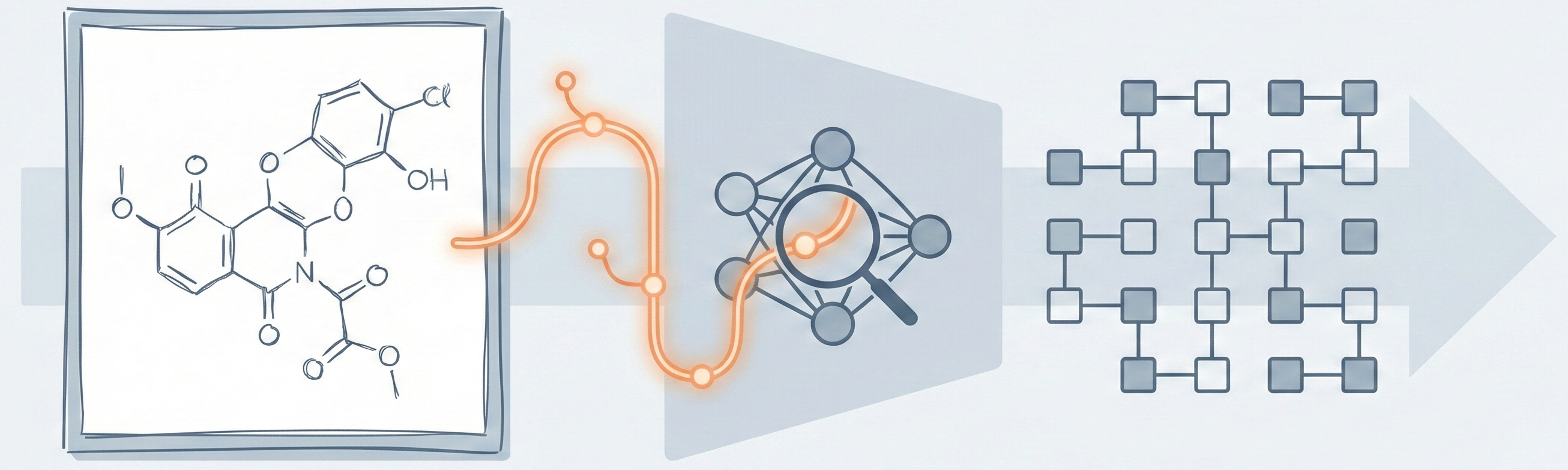
This is primarily a Method paper with a significant Resource component.
- Method: It proposes a novel architectural framework (RCGD) and a new representation syntax (SSML) to solve the specific problem of handwritten chemical structure recognition.
- Resource: It introduces a new benchmark dataset, EDU-CHEMC, containing 50,000 handwritten images to address the lack of public data in this domain.
What is the motivation?
Recognizing handwritten chemical structures is significantly harder than printed ones due to:
- Inherent Ambiguity: Handwritten atoms and bonds vary greatly in appearance.
- Projection Complexity: Converting 2D projected layouts (like Natta or Fischer projections) into linear strings is difficult.
- Limitations of Existing Formats: Standard formats like SMILES require domain knowledge (valence rules) and have a high semantic gap with the visual image. They often fail to represent “invalid” structures commonly found in educational/student work.
What is the novelty here?
The paper introduces two core contributions to bridge the semantic gap between image and markup:
Structure-Specific Markup Language (SSML): An extension of Chemfig that provides an unambiguous, visual-based graph representation. Unlike SMILES, it describes how to draw the molecule step-by-step, making it easier for models to learn visual alignments. It supports “reconnection marks” to handle cyclic structures explicitly.
Random Conditional Guided Decoder (RCGD): A decoder that treats recognition as a graph traversal problem. It introduces three novel mechanisms:
- Conditional Attention Guidance: Uses branch angle directions to guide the attention mechanism, preventing the model from getting lost in complex structures.
- Memory Classification: A module that explicitly stores and classifies “unexplored” branch points to handle ring closures (reconnections).
- Path Selection: A training strategy that randomly samples traversal paths to prevent overfitting to a specific serialization order.
What experiments were performed?
Datasets:
- Mini-CASIA-CSDB (Printed): A subset of 97,309 synthetic images, upscaled to $500 \x500$ resolution.
- EDU-CHEMC (Handwritten): A new dataset of 52,987 images collected from educational settings (cameras, scanners, screens), including erroneous/non-existent structures.
Baselines:
- Compared against standard String Decoders (SD) (based on DenseWAP) trained on SMILES strings.
- Compared against BTTR and ABM (recent mathematical expression recognition models) adapted for this task.
Ablation Studies:
- Evaluated the impact of removing Path Selection (PS) and Memory Classification (MC) mechanisms.
- Tested robustness to image rotation ($180^\circ$).
What were the outcomes and conclusions drawn?
- Superiority of SSML: Models trained with SSML significantly outperformed those trained with SMILES (92.09% vs 81.89% EM on printed data) due to reduced semantic gap.
- SOTA Performance: RCGD achieved the highest Exact Match (EM) scores on both datasets:
- Mini-CASIA-CSDB: 95.01% EM.
- EDU-CHEMC: 62.86% EM.
- Robustness: RCGD showed minimal performance drop (0.85%) on rotated images compared to SMILES-based methods (10.36% drop).
- Educational Utility: The method can recognize and reconstruct chemically invalid structures (e.g., a Carbon atom with 5 bonds), making it suitable for automated grading systems.
Reproducibility Details
Data
1. EDU-CHEMC (Handwritten)
- Total Size: 52,987 images.
- Splits: Training (48,998), Validation (999), Test (2,992).
- Characteristics: Real-world educational data, mixture of isolated molecules and reaction equations, includes invalid chemical structures.
2. Mini-CASIA-CSDB (Printed)
- Total Size: 97,309 images.
- Splits: Training (80,781), Validation (8,242), Test (8,286).
- Preprocessing: Original $300 \x300$ images were upscaled to $500 \x500$ RGB to resolve blurring issues.
Algorithms
1. SSML Generation
To convert a molecular graph to SSML:
- Traverse: Start from the left-most atom.
- Bonds/Atoms: Output atom text and bond format
<bond>[:<angle>]. - Branches: At branch points, use phantom symbols
(and)to enclose branches, ordered by ascending bond angle. - Reconnections: Use
?[tag]and?[tag, bond]to mark start/end of ring closures.
2. RCGD Specifics
- RCGD-SSML: Modified version of SSML for the decoder. Removes
()delimiters; adds\eob(end of branch). Maintains a dynamic Branch Angle Set ($M$). - Path Selection: During training, when multiple branches exist in $M$, the model randomly selects one to traverse next. During inference, it uses beam search to score candidate paths.
- Loss Function: $L_{\text{total}} = L_{\text{ce}} + L_{\text{bc}}$
- $L_{\text{ce}}$: Cross-entropy loss for character sequence generation.
- $L_{\text{bc}}$: Multi-label classification loss for the memory module (predicting reconnection bond types for stored branch states).
Models
Encoder: DenseNet
- Structure: 3 dense blocks.
- Growth Rate: 24.
- Depth: 32 per block.
- Output: High-dimensional feature map $x \in \mathbb{R}^{d_x \xh \xw}$.
Decoder: GRU with Attention
- Hidden State Dimension: 256.
- Embedding Dimension: 256.
- Attention Projection: 128.
- Memory Classification Projection: 256.
Training Config:
- Optimizer: Adam.
- Learning Rate: 2e-4 with multi-step decay (gamma 0.5).
- Dropout: 15%.
- Strategy: Teacher-forcing used for validation selection.
Evaluation
Metrics:
- Exact Match (EM): Percentage of samples where the predicted graph structure perfectly matches the label. For SMILES, string comparison; for SSML, converted to graph for isomorphism check.
- Structure EM: Auxiliary metric for samples with mixed content (text + molecules), counting samples where all molecular structures are correct.
Code Availability:
- The dataset is hosted at: https://github.com/iFLYTEK-CV/EDU-CHEMC
Citation
@inproceedings{huHandwrittenChemicalStructure2023,
title = {Handwritten Chemical Structure Image to Structure-Specific Markup Using Random Conditional Guided Decoder},
booktitle = {Proceedings of the 31st ACM International Conference on Multimedia},
author = {Hu, Jinshui and Wu, Hao and Chen, Mingjun and Liu, Chenyu and Wu, Jiajia and Yin, Shi and Yin, Baocai and Yin, Bing and Liu, Cong and Du, Jun and Dai, Lirong},
year = {2023},
month = oct,
pages = {8114--8124},
publisher = {ACM},
address = {Ottawa ON Canada},
doi = {10.1145/3581783.3612573},
isbn = {979-8-4007-0108-5}
}