Paper Information
Citation: Hong, C., Du, X., & Zhang, L. (2015). Research on Chemical Expression Images Recognition. Proceedings of the 2015 Joint International Mechanical, Electronic and Information Technology Conference, 267-271. https://doi.org/10.2991/jimet-15.2015.50
Publication: JIMET 2015 (Atlantis Press)
Additional Resources:
What kind of paper is this?
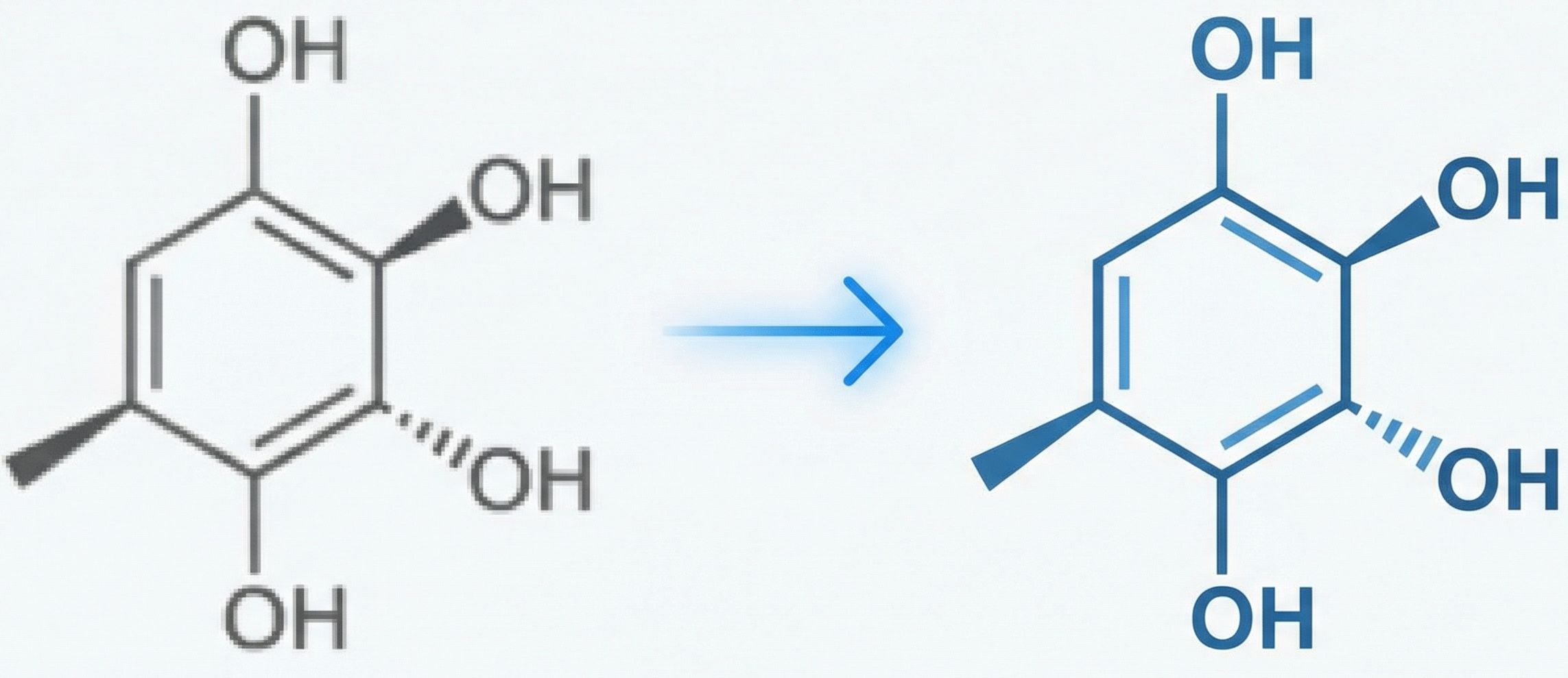
Method. The paper proposes a novel algorithmic pipeline (OCSR) for recognizing 2D organic chemical structures from images. It validates this method by comparing it against an existing tool (OSRA) using a quantitative metric (Tanimoto Coefficient) on a test set of 200 images.
What is the motivation?
A vast amount of chemical structural information exists in scientific literature (PDFs/images) that is not machine-readable. Manually converting these images to formats like InChI or CML is labor-intensive. Existing tools face challenges with:
- Adhesion: Poor separation when chemical symbols touch or overlap with bonds.
- Stereochemistry: Incomplete identification of “real” (solid) and “virtual” (dashed/hashed) wedge bonds.
What is the novelty here?
The authors propose a specific OCSR (Optical Chemical Structure Recognition) workflow with two key technical improvements:
- Vector-based Separation: The method vectorizes the image (using Potrace) to extract straight lines and curves, allowing better separation of “adhesive” chemical symbols (like H, N, O attached to bonds).
- Stereochemical Logic: Specific rules for identifying wedge bonds:
- Virtual (Dashed) Wedges: Identified by grouping connected domains and checking linear correlation of their center points.
- Real (Solid) Wedges: Identified after thinning by analyzing linear correlation and width variance of line segments.
What experiments were performed?
- Dataset: 200 chemical structure images collected from the network.
- Baselines: Compared against OSRA (Optical Structure Recognition Application), a free online tool.
- Metric: Tanimoto Coefficient, measuring the similarity of the set of recognized bonds and symbols against the ground truth.
What outcomes/conclusions?
- Performance: The proposed OCSR method achieved higher recognition rates than OSRA.
- Exact Match (100%): OCSR achieved 90.0% vs. OSRA’s 82.2%.
- High Similarity (>85%): OCSR recognized 157 structures vs. OSRA’s 114.
- Limitations: The paper notes that “real wedge” and “virtual wedge” identification was a primary focus, but general recognition effectiveness still “has room for improvement”.
Reproducibility Details
Data
The study used a custom collection of images, not a standard benchmark.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Evaluation | Web-crawled chemical images | 200 structures | Images containing 2D organic structures; specific source URLs not provided. |
Algorithms
The recognition pipeline follows these specific steps:
- Preprocessing:
- Grayscale: via
cvCvtColor(OpenCV). - Binarization: via Otsu’s method.
- Grayscale: via
- Isolated Symbol Removal:
- Identifies connected domains with aspect ratios in
[0.8, 3.0]. - Recognizes them using OCR (GOCR, OCRAD, Tesseract) and removes them from the image.
- Identifies connected domains with aspect ratios in
- Virtual Wedge Recognition:
- Groups small connected domains (points/clumps).
- Calculates linear correlation of center points; if collinear, treats as a dashed bond.
- Vectorization & Thinning:
- Thinning: Rosenfeld algorithm (optimized) to reduce lines to single pixel width.
- Vectorization: Uses Potrace to convert pixels to vector segments.
- Merging: Combines split vector segments based on angle thresholds to form long straight lines.
- Adhesive Symbol Separation:
- Identifies curves (short segments after vectorization) attached to long lines.
- Separates these domains and re-runs OCR.
- “Super Atom” Merging:
- Merges adjacent vertical/horizontal symbols (e.g., “HO”, “CH3”) based on distance thresholds between bounding boxes.
Models
The system relies on off-the-shelf OCR tools for character recognition; no custom ML models were trained.
- OCR Engines: GOCR, OCRAD, TESSERACT.
- Visualization: JSME (JavaScript Molecule Editor) used to render output strings.
Evaluation
| Metric | Value (OCSR) | Baseline (OSRA) | Notes |
|---|---|---|---|
| Exact Match (100%) | 90.0% | 82.2% | Percentage of 200 images perfectly recognized. |
| >95% Similarity | 95 images | 71 images | Count of images with Tanimoto > 0.95. |
| >85% Similarity | 157 images | 114 images | Count of images with Tanimoto > 0.85. |
Hardware
- Requirements: Unspecified; runs on standard CPU architecture (implied by use of standard libraries like OpenCV and Potrace).
Citation
@inproceedings{hongResearchChemicalExpression2015,
title = {Research on {{Chemical Expression Images Recognition}}},
booktitle = {Proceedings of the 2015 {{Joint International Mechanical}}, {{Electronic}} and {{Information Technology Conference}}},
author = {Hong, Chen and Du, Xiaoping and Zhang, Lu},
year = {2015},
publisher = {Atlantis Press},
address = {Chongqing, China},
doi = {10.2991/jimet-15.2015.50},
isbn = {978-94-6252-129-2}
}