Paper Information
Citation: Rajan, K., Brinkhaus, H. O., Agea, M. I., Zielesny, A., & Steinbeck, C. (2023). DECIMER.ai: an open platform for automated optical chemical structure identification, segmentation and recognition in scientific publications. Nature Communications, 14(1), 5045. https://doi.org/10.1038/s41467-023-40782-0
Publication: Nature Communications 2023
Additional Resources:
What kind of paper is this?
This is primarily a Resource paper (Infrastructure Basis) with a significant Method component.
The primary contribution is DECIMER.ai, a fully open-source platform (web app and Python packages) for the entire chemical structure mining pipeline, filling a gap where most tools were proprietary or fragmented. It also contributes the RanDepict toolkit for massive synthetic data generation.
The secondary methodological contribution proposes and validates a specific deep learning architecture (EfficientNet-V2 encoder + Transformer decoder) that treats chemical structure recognition as an image-to-text translation task (SMILES generation).
What is the motivation?
Data Scarcity: While the number of chemical publications is increasing, most chemical information is locked in non-machine-readable formats (images in PDFs) and is not available in public databases.
Limitations of Existing Tools: Prior OCSR (Optical Chemical Structure Recognition) tools were largely rule-based (fragile to noise) or proprietary.
Lack of Integration: There was no existing open-source system that combined segmentation (finding the molecule on a page), classification (confirming it is a molecule), and recognition (translating it to SMILES) into a single workflow.
What is the novelty here?
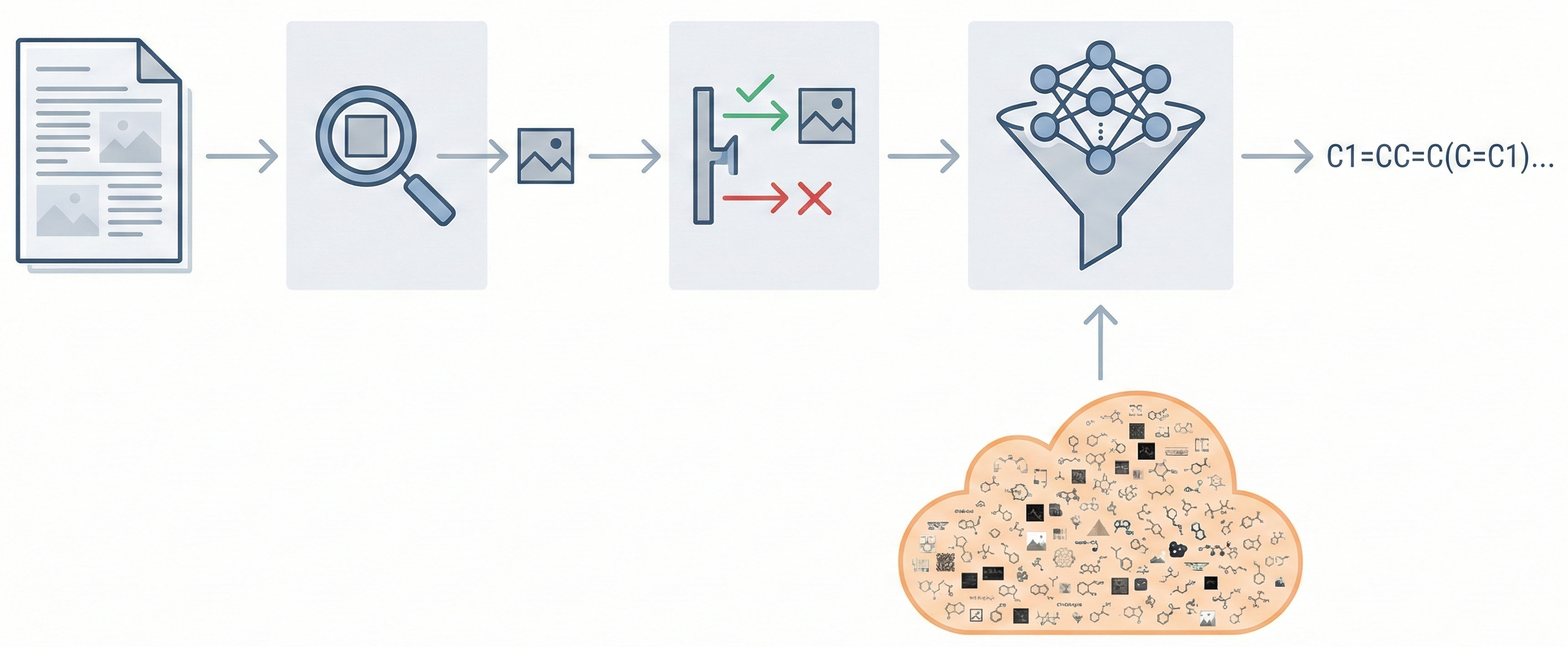
Comprehensive Workflow: It is the first open-source platform to integrate segmentation (Mask R-CNN), classification (EfficientNet), and recognition (Transformer) into a unified pipeline.
Data-Driven Approach: Unlike tools like MolScribe which use intermediate graph representations and rules, DECIMER uses a purely data-driven “image-to-SMILES” translation approach without hard-coded chemical rules.
Massive Synthetic Training: The use of RanDepict to generate over 450 million synthetic images, covering diverse depiction styles and augmentations (including Markush structures), to train the model from scratch.
What experiments were performed?
Benchmarking: The system was tested against openly available tools (OSRA, MolVec, Imago, Img2Mol, SwinOCSR, MolScribe) on standard datasets: USPTO, UOB, CLEF, JPO, and a custom “Hand-drawn” dataset.
Robustness Testing: Performance was evaluated on both clean images and images with added distortions (rotation, shearing) to test the fragility of rule-based systems vs. DECIMER.
Markush Structure Analysis: Specific evaluation of the model’s ability to interpret Markush structures (generic structures with R-groups).
Comparison of Approaches: A direct comparison with MolScribe by training DECIMER on MolScribe’s smaller training set to isolate the impact of architecture vs. data volume.
What were the outcomes and conclusions drawn?
SOTA Performance: DECIMER Image Transformer achieved the highest average Tanimoto similarity (>0.95) across all benchmarks and extremely low rates of catastrophic failure.
Data Volume Necessity: When trained on small datasets, MolScribe (graph/rule-based) outperformed DECIMER. DECIMER’s performance dominance relies on its massive training scale (>400M images).
Robustness: The model showed no performance degradation on distorted images, unlike rule-based legacy tools.
Generalization: Despite having no hand-drawn images in the training set, the model performed competitively on hand-drawn benchmarks, suggesting strong generalization capabilities.
Reproducibility Details
Data
The models were trained on synthetic data generated from PubChem molecules.
| Purpose | Dataset | Size | Generation/Notes |
|---|---|---|---|
| Training | pubchem_1 | ~100M mols | PubChem molecules (mass < 1500 Da), processed with RanDepict (v1.0.5). Included image augmentations. |
| Training | pubchem_2 | ~126M mols | Included Markush structures generated by pseudo-randomly replacing atoms with R-groups. Image size 299x299. |
| Training | pubchem_3 | >453M images | Re-depicted pubchem_2 molecules at 512x512 resolution. Used RanDepict v1.0.8. |
| Test | In-domain | 250,000 | Held-out set generated similarly to training data. |
| Benchmark | External | Various | USPTO (5719), UOB (5740), CLEF (992), JPO (450), Indigo (50k), Hand-drawn (5088). |
Data Generation:
- Tool: RanDepict (uses CDK, RDKit, Indigo, PIKACHU)
- Augmentations: Rotation, shearing, noise, pixelation, curved arrows, text labels
- Format: Data saved as TFRecord files for TPU training
Algorithms
- SMILES Tokenization: Regex-based splitting (atoms, brackets, bonds). Added
<start>,<end>, and padded with<pad>.<unk>used for unknown tokens. - Markush Token Handling: To avoid ambiguity, digits following ‘R’ (e.g., R1) were replaced with unique non-digit characters during training to distinguish them from ring-closure numbers.
- Image Augmentation Pipeline: Albumentations and custom RanDepict features were used to simulate “hand-drawn-like” styles.
Models
The platform consists of three distinct models:
DECIMER Segmentation:
- Architecture: Mask R-CNN (TensorFlow 2.10.0 implementation)
- Purpose: Detects and cuts chemical structures from full PDF pages
DECIMER Image Classifier:
- Architecture: EfficientNet-B0 (Noisy Student weights)
- Input: 224x224 pixels
- Training: Fine-tuned on ~10M images (balanced chemical/non-chemical)
- Performance: AUC 0.99 on in-domain test set
DECIMER Image Transformer (OCSR Engine):
- Encoder: EfficientNet-V2-M (CNN). Input size 512x512. 52M parameters
- Decoder: Transformer. 4 encoder blocks, 4 decoder blocks, 8 attention heads. d_model=512, d_ff=2048. 59M parameters
- Total Params: ~111 Million
Evaluation
- Primary Metric: Tanimoto Similarity (calculated on PubChem fingerprints of the predicted vs. ground truth SMILES)
- Secondary Metrics: Exact Match (Identity), BLEU score (for string similarity, esp. Markush)
- Failure Analysis: “Catastrophic failure” defined as Tanimoto similarity of 0 or invalid SMILES
Hardware
Training was performed on Google Cloud TPUs due to the massive dataset size.
pubchem_1/pubchem_2: Trained on TPU v3-32 pod slicepubchem_3(Final Model): Trained on TPU v3-256 pod slice- Training Time:
- Data generation (512x512): ~2 weeks on cluster (20 threads, 36 cores)
- Model Training (EffNet-V2-M): 1 day and 7 hours per epoch on TPU v3-256
Citation
@article{rajanDECIMERaiOpenPlatform2023,
title = {DECIMER.ai: an open platform for automated optical chemical structure identification, segmentation and recognition in scientific publications},
author = {Rajan, Kohulan and Brinkhaus, Henning Otto and Agea, M. Isabel and Zielesny, Achim and Steinbeck, Christoph},
journal = {Nature Communications},
volume = {14},
number = {1},
pages = {5045},
year = {2023},
doi = {10.1038/s41467-023-40782-0}
}