Paper Information
Citation: Rajan, K., Zielesny, A. & Steinbeck, C. (2020). DECIMER: towards deep learning for chemical image recognition. Journal of Cheminformatics, 12(65). https://doi.org/10.1186/s13321-020-00469-w
Publication: Journal of Cheminformatics 2020
Additional Resources:
What kind of paper is this?
This is primarily a Method ($\Psi_{\text{Method}}$) paper with a strong Resource ($\Psi_{\text{Resource}}$) component.
- Method: It proposes a novel architecture (DECIMER) that repurposes “show-and-tell” image captioning networks for Optical Chemical Entity Recognition (OCER), moving away from traditional rule-based segmentation.
- Resource: It establishes a framework for generating “potentially unlimited” synthetic training data using open-source cheminformatics tools (CDK) and databases (PubChem) to overcome the scarcity of annotated chemical images.
What is the motivation?
The extraction of chemical structures from scientific literature (OCER) is critical for populating open-access chemical databases. Traditional OCER systems (like OSRA or CLiDE) rely on complex, hand-tuned multi-step pipelines involving vectorization, character recognition, and graph compilation. These systems are brittle; incorporating new features is laborious. Inspired by AlphaGo Zero, the authors sought to replace these engineered rules with a deep learning approach that learns directly from massive amounts of data with minimal prior assumptions.
What is the novelty here?
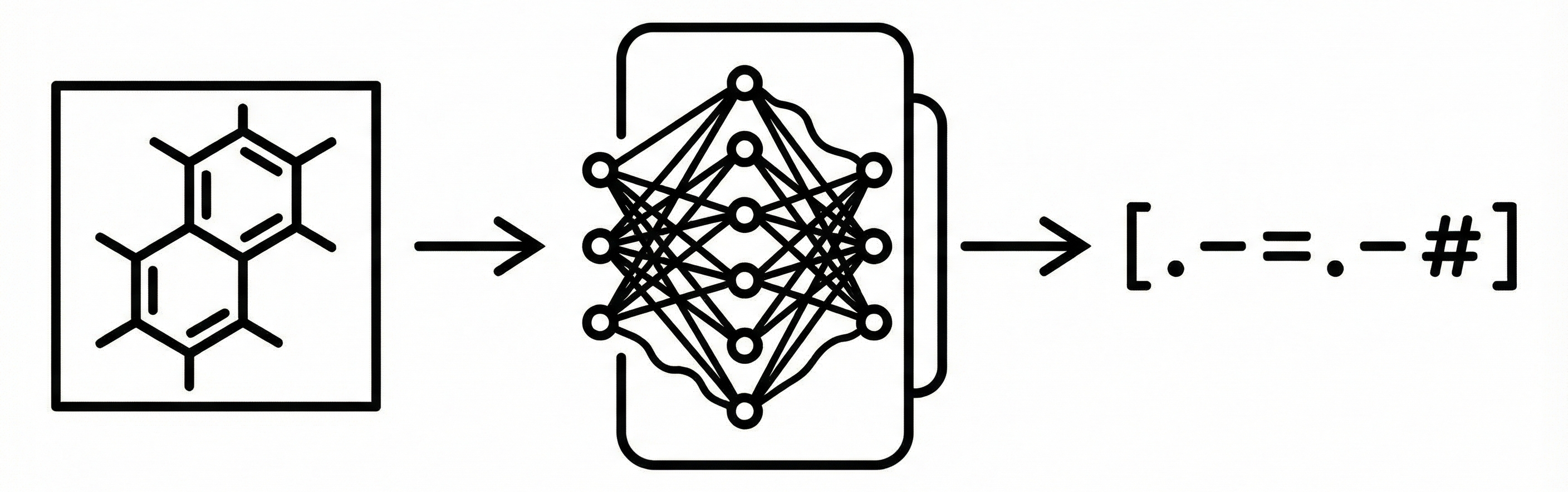
- Image-to-Text Formulation: The paper treats chemical structure recognition as a direct image captioning problem, translating a bitmap image directly into a SMILES string using an encoder-decoder network. This avoids explicit segmentation of atoms and bonds.
- Synthetic Data Strategy: The authors generate synthetic images from PubChem using the CDK Structure Diagram Generator, allowing for a dataset size that scales into the tens of millions.
- Robust String Representations: The study evaluates the impact of string representations on learning success, explicitly comparing standard SMILES against DeepSMILES and SELFIES to handle syntax validity more effectively.
What experiments were performed?
- Data Scaling: Models were trained on dataset sizes ranging from 54,000 to 15 million synthetic images to observe scaling laws regarding accuracy and training time.
- Representation Comparison: The authors compared the validity of predicted strings and recognition accuracy when training on SMILES versus DeepSMILES.
- Metric Evaluation: Performance was measured using:
- Tanimoto Similarity: Calculating the similarity between the fingerprint of the predicted molecule and the ground truth.
- Validity: The percentage of predicted strings that could be parsed into valid chemical graphs.
What outcomes/conclusions?
- Data Representation: DeepSMILES proved superior to standard SMILES for training stability and output validity. Preliminary tests suggested SELFIES performs even better (0.78 Tanimoto vs 0.53 for DeepSMILES at 6M images).
- Scaling Behavior: Accuracy improves linearly with dataset size. The authors extrapolate that near-perfect detection would require training on 50 to 100 million structures.
- Current Limitations: At the reported training scale (up to 15M), the model does not yet rival traditional rule-based approaches, but the learning curve suggests it is a viable path forward given sufficient compute and data.
Reproducibility Details
Data
The training data is synthetic, generated using the Chemistry Development Kit (CDK) Structure Diagram Generator (SDG) based on molecules from PubChem.
Curation Rules (applied to PubChem data):
- Molecular weight < 1500 Daltons.
- Elements restricted to: C, H, O, N, P, S, F, Cl, Br, I, Se, B.
- No counter ions or charged groups.
- No isotopes (e.g., D, T).
- Bond count between 5 and 40.
- SMILES length < 40 characters.
- Implicit hydrogens only (except in functional groups).
Preprocessing:
- Images: Generated as 299x299 bitmaps to match Inception V3 input requirements.
- Augmentation: One random rotation applied per molecule; no noise or blurring added in this iteration.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Training | Synthetic (PubChem) | 54k - 15M | Scaled across 12 experiments |
| Testing | Independent Set | 6k - 1.6M | 10% of training size |
Algorithms
- Architecture:
"Show, Attend and Tell"(Attention-based Image Captioning). - Optimization: Adam optimizer with learning rate 0.0005.
- Loss Function: Sparse Categorical Crossentropy.
- Training Loop: Trained for 25 epochs per model. Batch size of 640 images.
Models
The network is implemented in TensorFlow 2.0.
- Encoder: Inception V3 (Convolutional NN), used unaltered. Extracts feature vectors saved as NumPy arrays.
- Decoder: Gated Recurrent Unit (GRU) based Recurrent Neural Network (RNN) with soft attention mechanism.
- Embeddings: Image embedding dimension size of 600.
Evaluation
The primary metric is Tanimoto similarity (Jaccard index) on PubChem fingerprints, which is robust for measuring structural similarity even when exact identity is not reached.
| Metric | Definition |
|---|---|
| Tanimoto 1.0 | Percentage of predictions that are chemically identical to ground truth (isomorphic). |
| Average Tanimoto | Mean similarity score across the test set (captures partial correctness). |
| Validity | Percentage of predicted strings that are valid DeepSMILES/SMILES. |
Hardware
Training was performed on a single node.
- GPU: 1x NVIDIA Tesla V100.
- CPU: 2x Intel Xeon Gold 6230.
- RAM: 384 GB.
- Compute Time:
- Linear scaling with data size.
- 15 million structures took ~27 days (64,909s per epoch).
- Projected time for 100M structures: ~4 months on single GPU.
Citation
@article{rajanDECIMERDeepLearning2020,
title = {{{DECIMER}}: Towards Deep Learning for Chemical Image Recognition},
shorttitle = {{{DECIMER}}},
author = {Rajan, Kohulan and Zielesny, Achim and Steinbeck, Christoph},
year = 2020,
month = dec,
journal = {Journal of Cheminformatics},
volume = {12},
number = {1},
pages = {65},
issn = {1758-2946},
doi = {10.1186/s13321-020-00469-w}
}