Paper Information
Citation: Gkoutos, G. V., Rzepa, H., Clark, R. M., Adjei, O., & Johal, H. (2003). Chemical Machine Vision: Automated Extraction of Chemical Metadata from Raster Images. Journal of Chemical Information and Computer Sciences, 43(5), 1342-1355. https://doi.org/10.1021/ci034017n
Publication: J. Chem. Inf. Comput. Sci. 2003
What kind of paper is this?
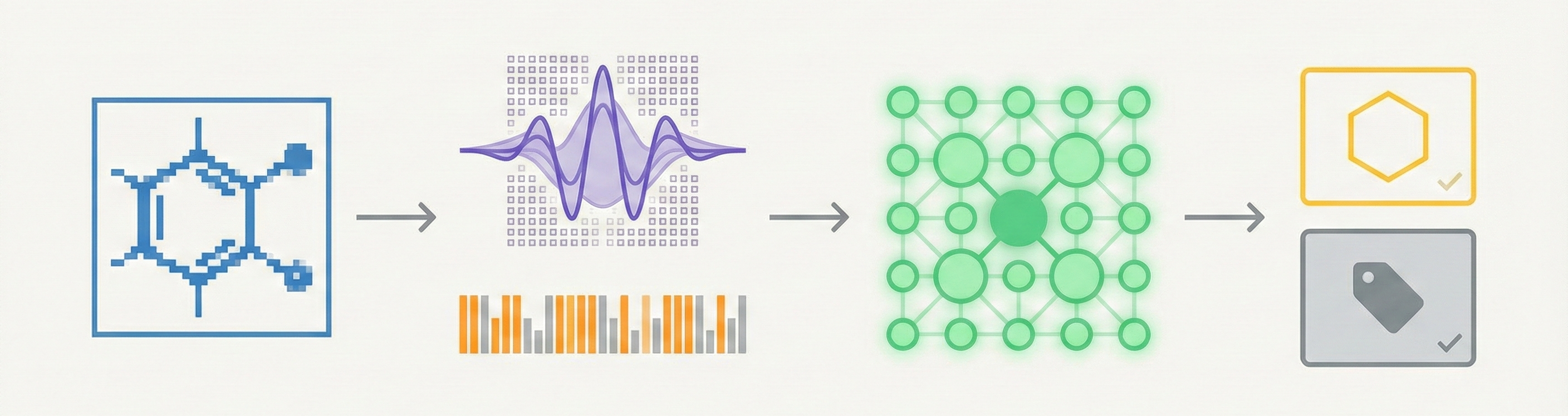
This is a Method paper. It proposes a novel architectural pipeline - specifically applying “machine vision” techniques (Gabor wavelets and Kohonen networks) - to the problem of identifying chemical diagrams in low-resolution raster images. The paper focuses on the “how” (the algorithm and its parameters) and validates the method through quantitative experiments optimizing feature vectors and masks.
What is the motivation?
The primary motivation is to unlock the “large amount of data” trapped in legacy raster images (GIF, JPEG) on the Web that lack semantic metadata.
- Legacy Data Problem: Most chemical structural information on the Web is embedded in raster images, not machine-readable formats like Molfiles.
- Limitations of Existing Tools: Previous tools like Kekule and CLiDE acted as “Chemical OCR,” attempting to reconstruct exact atom-bond connections. This required high-resolution images (>300 dpi) and human intervention, making them unsuitable for automated Web crawling of low-resolution (72-96 dpi) images.
- Goal: To create a low-cost, automated tool for a “robot-based Internet resource discovery tool” that can classify images (e.g., “is this a molecule?”).
What is the novelty here?
The core novelty is the shift from “Optical Character Recognition” (exact reconstruction) to “Texture Recognition” (classification).
- Texture-Based Approach: The authors treat chemical diagrams as textures. They use Gabor wavelets to extract texture features. Crucially, this system does not recognize specific chemical structures (i.e., atom-bond connectivity tables, SMILES, or Molfiles) - it only classifies images into broad categories.
- Incremental Learning: The system uses a Kohonen Self-Organizing Feature Map (KSOFM) combined with Class Boundary Analysis (CBA). This allows for “incremental learning,” where new classes (e.g., aromatic vs. non-aromatic) can be added without retraining the entire system.
- Optimization for Chemistry: The authors identify specific parameters (frequency channels, mask sizes) that are optimal for the “texture” of chemical diagrams.
- Integration with ChemDig: The method was designed to feed into ChemDig, a robot-based index engine for automated web crawling and metadata generation.
What experiments were performed?
The authors performed optimization and validation experiments using a dataset of 300 images divided into three classes: Ring Systems, Non-Ring Systems, and Non-Chemistry (textures, biological figures, etc.).
- Parameter Optimization: They systematically varied hyperparameters to find the optimal configuration:
- Feature Vector Size: Tested sizes from 100 to 4000 elements.
- Energy Mask Size: Tested windows from $3 \x3$ to $15 \x15$ pixels.
- Frequency Channels: Tested seven spatial frequencies ($\sqrt{2}$ to $64\sqrt{2}$).
- Classification Performance: Evaluated the system’s ability to classify unseen test images using a 50:50 training/test split.
- Comparison: Qualitatively compared the approach against vectorization tools (Autotrace, CR2V).
What were the outcomes and conclusions drawn?
- Optimal Configuration: The system performed best with a feature vector size of ~1500 elements, a $9 \x9$ energy mask, and frequency channel $4\sqrt{2}$.
- High Accuracy: Achieved a recognition rate of 91% with a 50:50 training/test split, and up to 92% with a 70:30 split.
- Robustness: The system successfully distinguished between chemical and non-chemical images (zero false negatives for chemical images).
- Limitations: Misclassifications occurred between “ring” and “non-ring” systems when structures had similar visual “textures” (e.g., similar density or layout).
- Impact: The method is viable for automating metadata generation (e.g.,
alttags) for web crawlers, functioning as a coarse-grained filter before more expensive processing.
Reproducibility Details
Data
The study used a custom dataset of raster images collected from the Web.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Training/Eval | Custom Web Dataset | 300 images | Split into 3 classes: Ring Systems, Non-Ring Systems, Non-Chemistry. |
| Resolution | Low-Res Web Images | 72-96 dpi | Deliberately chosen to mimic Web conditions where OCR fails. |
| Format | Raster | GIF, JPEG | Typical web formats. |
Algorithms
The core pipeline consists of a Gabor Transform Unit followed by a Training/Classification Unit.
- Gabor Wavelets: Used for feature extraction. The 2D Gabor wavelet equation is:
$$h(x,y)=exp{-\frac{1}{2}[\frac{x^{2}}{\sigma_{x}^{2}}+\frac{y^{2}}{\sigma_{y}^{2}}]}cos(2\pi\mu_{\sigma}x+\phi)$$
- Bank Structure: 28 filters total (4 orientations $\times$ 7 radial frequencies).
- Orientations: $0^{\circ}, 45^{\circ}, 90^{\circ}, 135^{\circ}$.
- Frequencies: 1 octave apart, specifically $1\sqrt{2}, \dots, 64\sqrt{2}$.
- Selected Frequency: $4\sqrt{2}$ was found to be optimal for chemistry.
- Preprocessing:
- Buffer Mounting: Images are mounted in a buffer (set to 0) to handle edge artifacts.
- Look-Up-Tables (LUT/LUF): A binary Look-Up-Frame (LUF) indicates Regions of Interest (ROI) to avoid computing empty space; values are stored in a Look-Up-Table (LUT) to prevent re-computation of overlapping windows.
- Feature Extraction:
- Non-linear Thresholding: $\psi(t) = \tanh(\alpha t)$ with $\alpha = 0.25$.
- Energy Function: Calculated as average absolute deviation from the mean using a window $W_{xy}$. $$e_{k}(x,y)=\frac{1}{M^{2}}\sum_{(a,b)\in Wxy}|\psi(r_{k}(a,b))|$$
- Optimal Window: $9 \x9$ pixels.
Models
The classification model relies on competitive learning.
- Architecture: Kohonen Self-Organizing Feature Map (KSOFM).
- Training:
- Learning Rate: Starts at 1.0, decreases to 0.1.
- Class Boundary Analysis (CBA): Computes the centroid (mean) and variance of each cluster. The variance defines the class boundary.
- Classification Metric: Euclidean Distance Norm. An unknown vector is classified based on the shortest distance to a cluster center, provided it falls within the variance boundary. $$D_{ij}=||x_{i}-x_{j}||$$
Evaluation
Performance was measured using recognition rate ($R_s$) and misclassification error ($E_s$).
| Metric | Value | Baseline | Notes |
|---|---|---|---|
| Recognition Rate | 91% | N/A | Achieved with 50:50 split. 92% with 70:30 split. |
| Feature Size | ~1500 | 4000 | Reducing vector size from 4000 to 1500 maintained ~80% accuracy while improving speed. |
Citation
@article{gkoutosChemicalMachineVision2003,
title = {Chemical {{Machine Vision}}: {{Automated Extraction}} of {{Chemical Metadata}} from {{Raster Images}}},
shorttitle = {Chemical {{Machine Vision}}},
author = {Gkoutos, Georgios V. and Rzepa, Henry and Clark, Richard M. and Adjei, Osei and Johal, Harpal},
year = 2003,
month = sep,
journal = {Journal of Chemical Information and Computer Sciences},
volume = {43},
number = {5},
pages = {1342--1355},
issn = {0095-2338},
doi = {10.1021/ci034017n},
urldate = {2025-12-15},
langid = {english}
}