Paper Information
Citation: Chang, M., Han, S., & Zhang, D. (2009). A Unified Framework for Recognizing Handwritten Chemical Expressions. 2009 10th International Conference on Document Analysis and Recognition, 1345–1349. https://doi.org/10.1109/ICDAR.2009.64
Publication: ICDAR 2009
What kind of paper is this?
This is a Methodological Paper ($\Psi_{\text{Method}}$). The authors propose a “unified framework” consisting of novel statistical algorithms for symbol grouping and structure analysis. The paper focuses on the architectural innovation required to handle the complexity of organic chemistry structures alongside inorganic formulas. It validates this method through ablation studies (grouping vs. structure vs. verification) and accuracy metrics on a specific dataset, fitting the “How well does this work?” core question of methodological papers.
What is the motivation?
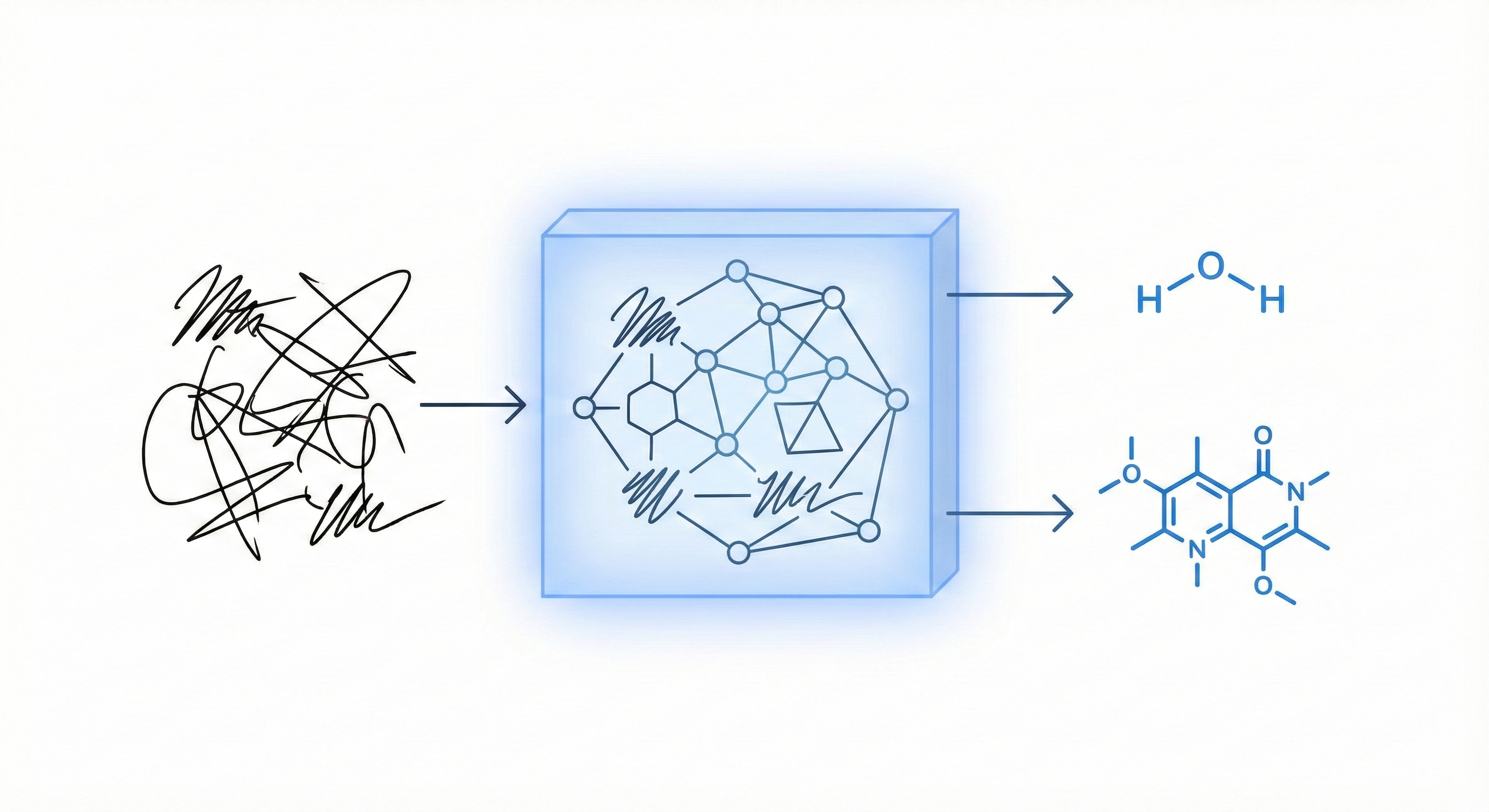
Handwritten scientific expression recognition is crucial for natural user interfaces in education. While math expression recognition had seen commercial progress by 2009, chemical expression recognition was less active.
The specific gap addressed is the complexity of organic chemical expressions. Unlike inorganic formulas (which resemble linear math equations), organic formulas have complex 2D structures (diagram-like) with various bond types and rings. Existing work often relied on strong assumptions (e.g., single-stroke symbols) or failed to handle arbitrary compounds. There was a practical need for a unified solution that could handle both inorganic and organic domains consistently.
What is the novelty here?
The core contribution is a unified statistical framework that treats inorganic and organic expressions within the same pipeline. Key technical novelties include:
- Unified Bond Modeling: A “bond modeling” approach where bonds are treated as special symbols. It introduces “extended bond symbols” (multi-stroke bonds) that are detected and then split into single/double/triple bonds using corner detection, allowing consistent processing.
- Chemical Expression Structure Graph (CESG): A defined graph representation for generic chemical expressions (nodes = symbols, edges = bonds/spatial relations).
- Non-Symbol Modeling: In the symbol grouping phase, the system explicitly models “non-symbols” (invalid groups) to reduce over-grouping errors.
- Global Graph Search: Structure analysis is formulated as finding the optimal CESG by searching over a “Weighted Direction Graph” ($G_{WD}$).
What experiments were performed?
The authors validated the framework on a proprietary database of 35,932 handwritten chemical expressions collected from 300 writers.
- Setup: The data was split into ~26k training and ~6.4k testing samples.
- Metric: Recognition accuracy was measured by expression (strict metric: all symbols + structure must be correct).
- Ablations: They evaluated the performance contribution of each component:
- Symbol Grouping alone.
- Symbol Grouping + Structure Analysis.
- Full system (Grouping + Structure + Semantic Verification).
What were the outcomes and conclusions drawn?
- Accuracy: The full framework achieved a Top-1 accuracy of 75.4% and a Top-5 accuracy of 83.1%.
- Component Contribution:
- Structure Analysis is the bottleneck; adding it drops theoretical “perfect grouping” performance from 85.9% to 74.1% (Top-1) due to structural errors.
- Semantic Verification (checking valence/grammar) improved relative accuracy by 1.7%.
- Conclusion: The unified framework effectively handles the variance in 2D space for chemical expressions, and the delayed decision-making (keeping top-N candidates) is effective.
Reproducibility Details
Data
The study used a private Microsoft Research Asia dataset, making direct reproduction difficult.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Total | Proprietary MSRA DB | 35,932 expressions | Written by 300 people |
| Training | Subset | 25,934 expressions | |
| Testing | Subset | 6,398 expressions |
- Content: 2,000 unique expressions from high school/college textbooks.
- Composition: ~25% of samples are organic expressions.
- Vocabulary: 163 symbol classes (elements, digits,
+,↑,%, bonds, etc.).
Algorithms
1. Symbol Grouping (Dynamic Programming)
- Objective: Find optimal symbol sequence $G_{max}$ maximizing $P(G|Ink)$.
- Non-symbol modeling: Iteratively trained models on “incorrect grouping results” to learn to reject invalid strokes.
- Inter-group modeling: Uses Gaussian Mixture Models (GMM) to model spatial relations ($R_j$) between groups.
2. Bond Processing
- Extended Bond Symbol: Recognizes connected strokes (e.g., a messy double bond written in one stroke) as a single “extended” symbol.
- Splitting: Uses Curvature Scale Space (CSS) corner detection to split extended symbols into primitive lines.
- Classification: A Neural Network verifies if the split lines form valid single, double, or triple bonds.
3. Structure Analysis (Graph Search)
- Graph Construction: Builds a Weighted Direction Graph ($G_{WD}$) where nodes are symbol candidates and edges are potential relationships ($E_{c}, E_{nc}, E_{peer}, E_{sub}$).
- Edge Weights: Calculated via Eq (5): $P(O|S) \xP(\text{Spatial}|R) \xP(\text{Context}|S,R)$.
- Spatial probability uses rectangular control regions and distance functions.
- Contextual probability uses statistical co-occurrence (e.g., ‘C’ often appears with ‘H’).
- Search: Breadth-first search with pruning to find the top-N optimal CESGs.
Models
- Symbol Recognition: Implementation details not specified, but likely HMM or NN based on the era. Bond verification explicitly uses a Neural Network.
- Spatial Models: Gaussian Mixture Models (GMM) are used to model the 9 spatial relations (e.g., Left-super, Above, Subscript).
- Semantic Model: A Context-Free Grammar (CFG) parser is used for final verification (e.g., ensuring digits aren’t reactants).
Evaluation
Evaluation is performed using “Expression-level accuracy”.
| Metric | Value (Top-1) | Value (Top-5) | Notes |
|---|---|---|---|
| Full Framework | 75.4% | 83.1% | |
| Without Semantics | 74.1% | 83.0% | |
| Grouping Only | 85.9% | 95.6% | Theoretical max if structure analysis was perfect |
Citation
@inproceedings{changUnifiedFrameworkRecognizing2009,
title = {A {{Unified Framework}} for {{Recognizing Handwritten Chemical Expressions}}},
booktitle = {2009 10th {{International Conference}} on {{Document Analysis}} and {{Recognition}}},
author = {Chang, Ming and Han, Shi and Zhang, Dongmei},
year = 2009,
pages = {1345--1349},
publisher = {IEEE},
address = {Barcelona, Spain},
doi = {10.1109/ICDAR.2009.64}
}