Paper Information
Citation: Oldenhof, M., De Brouwer, E., Arany, A., & Moreau, Y. (2024). Atom-Level Optical Chemical Structure Recognition with Limited Supervision. arXiv preprint arXiv:2404.01743. https://doi.org/10.48550/arXiv.2404.01743
Publication: arXiv 2024
Additional Resources:
What kind of paper is this?
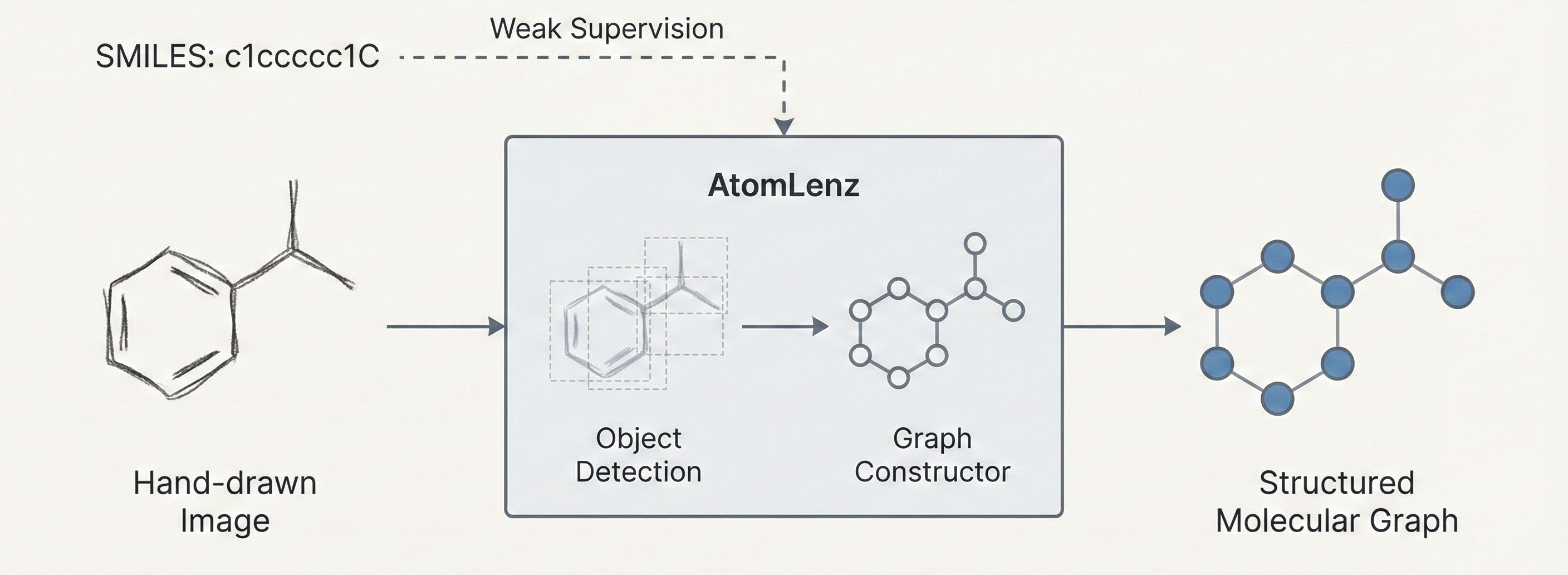
Methodological Paper with a strong Resource component.
The paper proposes a novel architecture (AtomLenz) and training framework (ProbKT* + Edit-Correction) to solve the problem of Optical Chemical Structure Recognition (OCSR) in data-sparse domains. It also releases a curated, relabeled dataset of hand-drawn molecules to support this task.
What is the motivation?
Optical Chemical Structure Recognition (OCSR) is critical for digitizing chemical literature and lab notes. However, existing methods face two main limitations:
- Generalization: They struggle with sparse or stylistically unique domains, such as hand-drawn images.
- Annotation Cost: “Atom-level” methods (which detect individual atoms/bonds) require expensive bounding box annotations, which are rarely available for real-world data like hand sketches.
- Lack of Localization: “Image-to-SMILES” methods (like DECIMER) work well but fail to provide interpretability or localization of atoms in the original image.
What is the novelty here?
The core contribution is AtomLenz, the first OCSR framework to achieve atom-level entity detection using only SMILES supervision.
- Weakly Supervised Training: They employ ProbKT* (Probabilistic Knowledge Transfer), a logic-based reasoning module that allows the object detector to learn from SMILES strings without explicit bounding boxes.
- Edit-Correction Mechanism: A strategy that generates pseudo-labels by finding the smallest graph edit distance between the predicted graph and the ground truth SMILES, enabling fine-tuning on rare atom types.
- ChemExpert: A chemically informed ensemble strategy that cascades predictions from multiple models (e.g., AtomLenz + DECIMER), validating them against chemical rules to select the best output.
What experiments were performed?
The authors evaluated the model on domain adaptation and data efficiency:
- Pretraining: Trained on ~214k synthetic images from ChEMBL (generated via RDKit).
- Target Domain: Fine-tuned on the Brinkhaus hand-drawn dataset (4,070 images) using only SMILES supervision.
- Evaluation Sets:
- Hand-drawn test set: 1,018 images
- ChemPix: 614 out-of-domain hand-drawn images
- Atom Localization set: 1,000 synthetic images with bbox annotations
- Baselines: Compared against DECIMER (v2.2), Img2Mol, MolScribe, ChemGrapher, and OSRA.
What were the outcomes and conclusions drawn?
- SOTA Performance: The ensemble ChemExpert (combining AtomLenz and DECIMER) achieved state-of-the-art accuracy on both hand-drawn (63.5%) and ChemPix (51.8%) test sets.
- Effective Weak Supervision: The EditKT* strategy significantly boosted performance over the base model (from 0.9% to 33.8% accuracy on hand-drawn data), proving the viability of learning from SMILES alone.
- Data Efficiency: AtomLenz outperformed baselines when trained from scratch on small datasets (4k images), demonstrating superior sample efficiency compared to Transformer-based methods like DECIMER which require millions of samples.
- Localization: The model provides accurate bounding boxes (mAP 0.801), unlike pure image-to-SMILES models.
Reproducibility Details
Data
The study utilizes a mix of large synthetic datasets and smaller curated hand-drawn datasets.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Pretraining | Synthetic ChEMBL | ~214,000 | Generated via RDKit/Indigo. Annotated with atoms, bonds, charges, stereocenters. |
| Fine-tuning | Hand-drawn (Brinkhaus) | 4,070 | Used for weakly supervised adaptation (SMILES only). |
| Evaluation | Hand-drawn Test | 1,018 | |
| Evaluation | ChemPix | 614 | Out-of-distribution hand-drawn images. |
| Evaluation | Atom Localization | 1,000 | Synthetic images with ground truth bounding boxes. |
Algorithms
- Molecular Graph Constructor (Algorithm 1): A rule-based system to assemble the graph from detected objects:
- Filtering: Removes overlapping atom boxes (IoU threshold).
- Node Creation: Merges charge and stereocenter objects with the nearest atom objects.
- Edge Creation: Iterates over bond objects; if a bond overlaps with exactly two atoms, an edge is added. If >2, it selects the most probable pair.
- Validation: Checks valency constraints; removes bonds iteratively if constraints are violated.
- Weakly Supervised Training:
- ProbKT*: Uses Hungarian matching to align predicted objects with the “ground truth” implied by the SMILES string, allowing backpropagation without explicit boxes.
- Graph Edit-Correction: Computes the Minimum Graph Edit Distance between the predicted graph and the true SMILES graph to generate pseudo-labels for retraining.
Models
- Object Detection Backbone: Faster R-CNN.
- Four distinct models are trained for different entity types: Atoms ($O^a$), Bonds ($O^b$), Charges ($O^c$), and Stereocenters ($O^s$).
- Loss Function: Multi-task loss combining Multi-class Log Loss ($L_{cls}$) and Regression Loss ($L_{reg}$).
- ChemExpert: An ensemble wrapper that prioritizes models based on user preference (e.g., DECIMER first, then AtomLenz). It accepts the first prediction that passes RDKit chemical validity checks.
Evaluation
Primary metrics focused on structural correctness and localization accuracy.
| Metric | Value (Hand-drawn) | Baseline (DECIMER FT) | Notes |
|---|---|---|---|
| Accuracy (T=1) | 33.8% (AtomLenz+EditKT*) | 62.2% | Exact ECFP6 fingerprint match. |
| Tanimoto Sim. | 0.484 | 0.727 | Average similarity. |
| mAP | 0.801 | N/A | Localization accuracy (IoU 0.05-0.35). |
| Ensemble Acc. | 63.5% | 62.2% | ChemExpert (DECIMER + AtomLenz). |
Hardware
- Compute: Experiments utilized the Flemish Supercomputer Center (VSC) resources.
- Note: Specific GPU models (e.g., A100/V100) are not explicitly detailed in the text, but Faster R-CNN training is standard on consumer or enterprise GPUs.
Citation
@misc{oldenhofAtomLevelOpticalChemical2024,
title = {Atom-{{Level Optical Chemical Structure Recognition}} with {{Limited Supervision}}},
author = {Oldenhof, Martijn and Brouwer, Edward De and Arany, Adam and Moreau, Yves},
year = 2024,
month = apr,
number = {arXiv:2404.01743},
eprint = {2404.01743},
primaryclass = {cs},
publisher = {arXiv},
doi = {10.48550/arXiv.2404.01743},
urldate = {2025-10-25},
abstract = {Identifying the chemical structure from a graphical representation, or image, of a molecule is a challenging pattern recognition task that would greatly benefit drug development. Yet, existing methods for chemical structure recognition do not typically generalize well, and show diminished effectiveness when confronted with domains where data is sparse, or costly to generate, such as hand-drawn molecule images. To address this limitation, we propose a new chemical structure recognition tool that delivers state-of-the-art performance and can adapt to new domains with a limited number of data samples and supervision. Unlike previous approaches, our method provides atom-level localization, and can therefore segment the image into the different atoms and bonds. Our model is the first model to perform OCSR with atom-level entity detection with only SMILES supervision. Through rigorous and extensive benchmarking, we demonstrate the preeminence of our chemical structure recognition approach in terms of data efficiency, accuracy, and atom-level entity prediction.},
archiveprefix = {arXiv},
langid = {english},
keywords = {Computer Science - Computer Vision and Pattern Recognition},
file = {/Users/hunterheidenreich/Zotero/storage/4ILTDIFX/Oldenhof et al. - 2024 - Atom-Level Optical Chemical Structure Recognition with Limited Supervision.pdf}
}