Paper Information
Citation: Algorri, M.-E., Zimmermann, M., Friedrich, C. M., Akle, S., & Hofmann-Apitius, M. (2007). Reconstruction of Chemical Molecules from Images. Proceedings of the 29th Annual International Conference of the IEEE EMBS, 4609–4612. https://doi.org/10.1109/IEMBS.2007.4353366
Publication: IEEE EMBS 2007
What kind of paper is this?
$\Psi_{\text{Method}}$ (Methodological Basis)
This paper is a clear methodological contribution describing a novel system architecture. It proposes a five-stage pipeline to solve a specific engineering problem: converting rasterized chemical images into structured chemical files (SDF). The authors validate the method by benchmarking it against a commercial product (CLIDE) and analyzing performance across multiple databases.
What is the motivation?
- Data Inaccessibility: A massive amount of chemical knowledge (scientific articles, patents) exists only as raster images, rendering it inaccessible to computational analysis.
- Inefficiency of Manual Entry: Manual replication of molecules into CAD programs is the standard but unscalable solution for extracting this information.
- Limitations of Existing Tools: Previous academic and commercial attempts (early 90s systems like CLIDE) had faded or remained limited in robustness, leaving the problem “wide open”.
What is the novelty here?
The core novelty is the topology-preserving vectorization strategy designed specifically for chemical graphs rather than general engineering drawings.
- Graph-Centric Vectorizer: Unlike CAD vectorizers that prioritize pixel precision, this system prioritizes graph characteristics—ensuring one line in the image becomes exactly one vector, regardless of line width or vertex thickness.
- Chemical Knowledge Module: The inclusion of a final validation step that applies chemical rules (valence, charge) to detect and potentially correct reconstruction errors.
- Hybrid Recognition: The separation of the pipeline into a “Body” path (vectorizer for bonds) and an “OCR” path (SVM for atomic symbols), which are re-integrated in a reconstruction phase.
What experiments were performed?
The authors performed a quantitative validation using ground-truth SDF files to verify reconstruction accuracy.
- Baselines: The system was benchmarked against the commercial software CLIDE on “Database 1”.
- Datasets: Three distinct databases were used:
- Database 1: 100 images (varied fonts/line widths).
- Database 2: 100 images.
- Database 3: 7,604 images (large-scale test).
What were the outcomes and conclusions drawn?
- Superior Performance: On Database 1, the proposed system correctly reconstructed 97% of images, whereas the commercial CLIDE system only reconstructed 25% (after parameter tuning).
- Scalability: The system maintained reasonable performance on the large dataset (Database 3), achieving 67% accuracy.
- Robustness: The system can handle varying fonts and line widths via parameterization.
- Future Work: The authors plan to implement a feedback loop where the Chemical Knowledge Module can send error signals back to earlier modules to correct inconsistencies.
Reproducibility Details
Data
The paper utilizes three databases for validation. The authors note that for these images, the correct SDF files were already available, allowing for direct automated checking.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Evaluation | Database 1 | 100 Images | Varied line widths, fonts, symbols; used for CLIDE comparison. |
| Evaluation | Database 2 | 100 Images | General chemical database. |
| Evaluation | Database 3 | 7,604 Images | Large-scale database. |
Algorithms
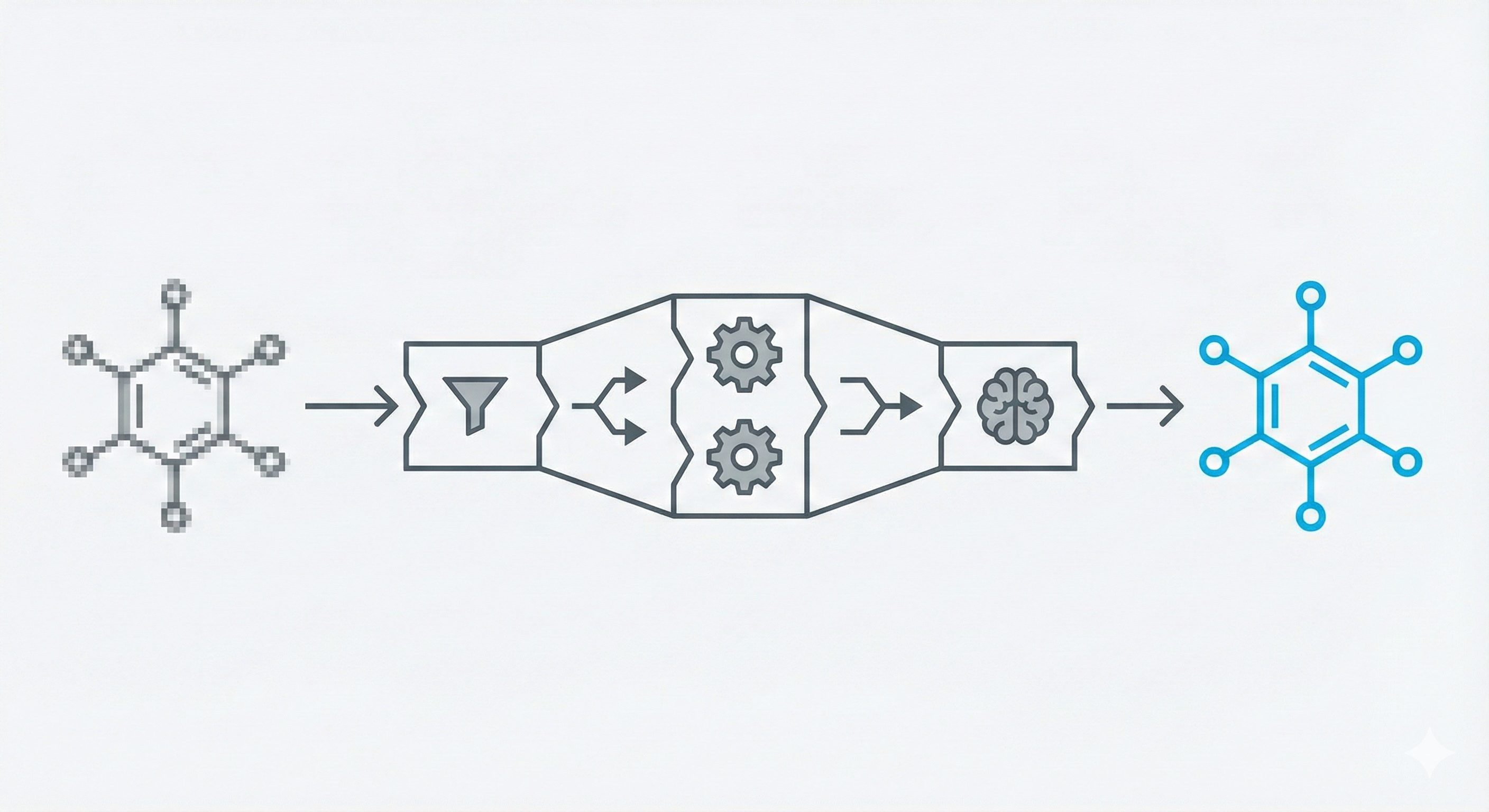
The system is composed of five distinct modules executed in sequence:
1. Binarization & Segmentation
- Preprocessing: Removal of anti-aliasing effects followed by adaptive histogram binarization.
- Connected Components: A non-recursive raster-scan algorithm identifies connected Run-Length Encoded (RLE) segments.
2. Optical Character Recognition (OCR)
- Feature Extraction: Uses functions similar to Zernike moments and a wavelet transform strategy.
- Classification: Identifies isolated characters/symbols and separates them from the molecular “body”.
3. Vectorizer
- Logic: Assigns local directions to RLE segments based on neighbors, then groups segments with similar local direction patterns.
- Constraint: Enforces a 1-to-1 mapping between visual lines and graph vectors to prevent spurious small vectors at thick joints.
4. Reconstruction (Heuristics)
This module annotates vectors with chemical significance:
- Chiral Bonds (Wedges): Identified by registering vectors against original pixel density. If a vector corresponds to a thick geometric form (triangle/rectangle), it is labeled chiral.
- Dotted Chiral Bonds: Identified by clustering isolated vectors (no neighbors) using quadtree clustering on geometric centers. Coherent parallel clusters are fused into a single bond.
- Double/Triple Bonds: Detected by checking for parallel vectors within a Region of Interest (ROI) defined as the vector’s bounding box dilated by a factor of 2.
- Superatoms: OCR results are clustered by dilating bounding boxes; overlapping boxes are grouped into names (e.g., “COOH”).
5. Chemical Knowledge
Validates the generated graph against rules for valences and charges. If valid, an SDF file is generated.
Models
- SVM (Support Vector Machine): Used within the OCR module to classify connected components as characters or symbols. It is trained to be tolerant to rotation and font variations.
Evaluation
The primary metric is binary success rate per molecule (perfect reconstruction of the SDF).
| Metric | Value (DB1) | Value (DB3) | Baseline (CLIDE on DB1) | Notes |
|---|---|---|---|---|
| Correct Reconstruction | 97% | 67% | 25% | CLIDE required significant parameter tuning to reach 25%. |
Citation
@inproceedings{algorriReconstructionChemicalMolecules2007,
title = {Reconstruction of {{Chemical Molecules}} from {{Images}}},
booktitle = {Proceedings of the 29th Annual International Conference of the IEEE EMBS},
author = {Algorri, Maria-Elena and Zimmermann, Marc and Friedrich, Christoph M. and Akle, Santiago and {Hofmann-Apitius}, Martin},
year = {2007},
pages = {4609--4612},
publisher = {IEEE},
doi = {10.1109/IEMBS.2007.4353366}
}