Paper Information
Citation: Zhang, X.-C., Yi, J.-C., Yang, G.-P., Wu, C.-K., Hou, T.-J., & Cao, D.-S. (2022). ABC-Net: A divide-and-conquer based deep learning architecture for SMILES recognition from molecular images. Briefings in Bioinformatics, 23(2), bbac033. https://doi.org/10.1093/bib/bbac033
Publication: Briefings in Bioinformatics 2022
Additional Resources:
What kind of paper is this?
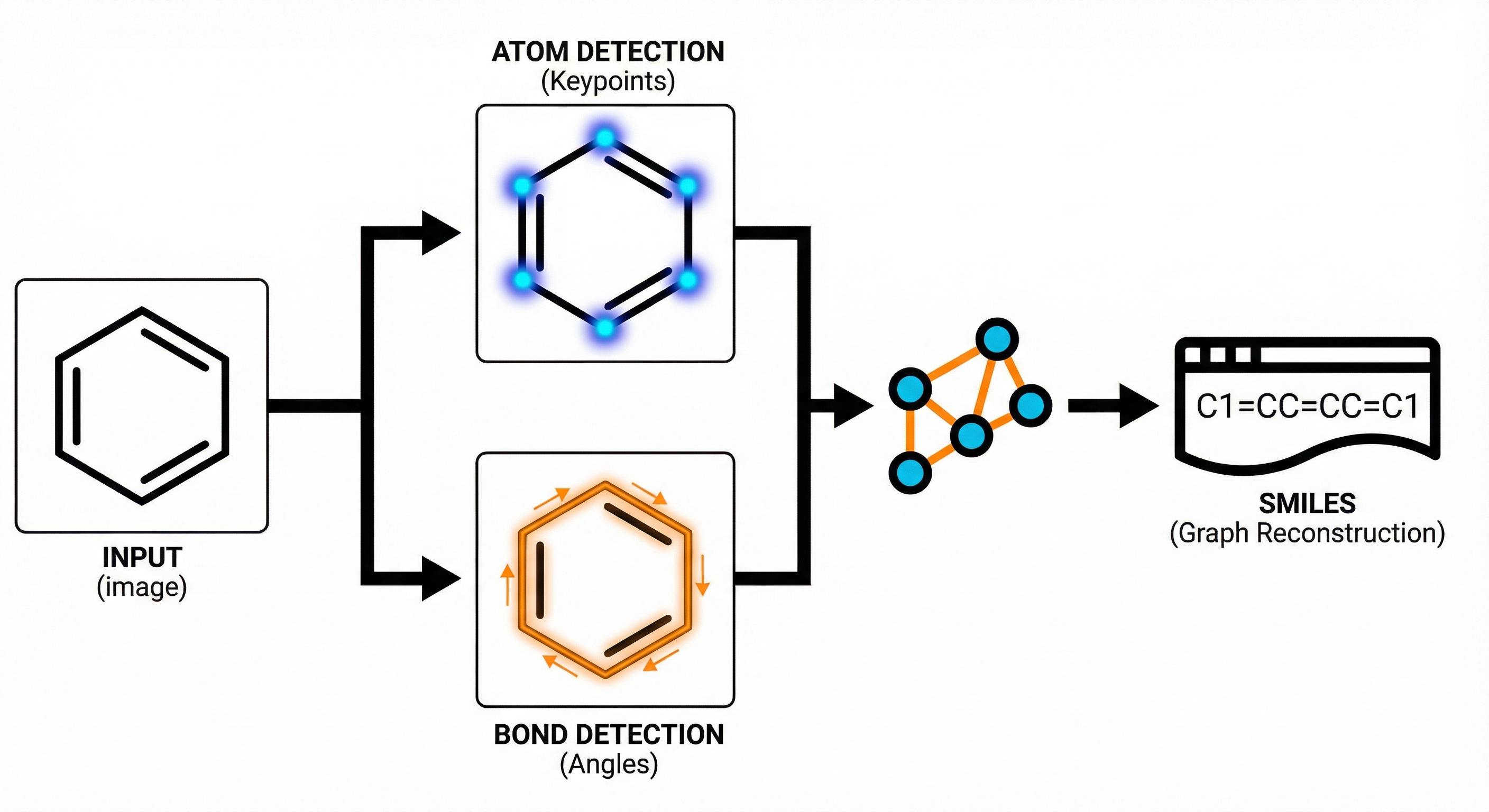
Method. The paper proposes a novel architectural framework (ABC-Net) for Optical Chemical Structure Recognition (OCSR). It reformulates the problem from image captioning (sequence generation) to keypoint estimation (pixel-wise detection), backed by ablation studies on noise and comparative benchmarks against SOTA tools.
What is the motivation?
- Inefficiency of Rule-Based Methods: Traditional tools (OSRA, MolVec) rely on hand-coded rules that are brittle, require domain expertise, and fail to handle the wide variance in molecular drawing styles.
- Data Inefficiency of Captioning Models: Recent Deep Learning approaches (like DECIMER, Img2mol) treat OCSR as image captioning (Image-to-SMILES). This is data-inefficient because canonical SMILES require learning traversal orders, necessitating millions of training examples.
- Goal: To create a scalable, data-efficient model that predicts graph structures directly by detecting atomic/bond primitives.
What is the novelty here?
- Divide-and-Conquer Strategy: ABC-Net breaks the problem down into detecting atom centers and bond centers as independent keypoints.
- Keypoint Estimation: It leverages a Fully Convolutional Network (FCN) to generate heatmaps for object centers. This is inspired by computer vision techniques like CornerNet and CenterNet.
- Angle-Based Bond Detection: To handle overlapping bonds, the model classifies bond angles into 60 distinct bins ($0-360°$) at detected bond centers, allowing separation of intersecting bonds.
- Implicit Hydrogen Prediction: The model explicitly predicts the number of implicit hydrogens for aromatic heteroatoms to resolve ambiguity in dearomatization.
What experiments were performed?
- Dataset Construction: Synthetic dataset of 100,000 molecules from ChEMBL, rendered using two different engines (RDKit and Indigo) to ensure style diversity.
- Baselines: Compared against two rule-based methods (MolVec, OSRA) and one deep learning method (Img2mol).
- Robustness Testing: Evaluated on the external UOB dataset (real-world images) and synthetic images with varying levels of salt-and-pepper noise (up to $p=0.6$).
- Data Efficiency: Analyzed performance scaling with training set size (10k to 160k images).
What were the outcomes and conclusions drawn?
- Superior Accuracy: ABC-Net achieved 94-98% accuracy on synthetic test sets, significantly outperforming MolVec (<50%), OSRA (~61%), and Img2mol (~89-93%).
- Generalization: On the external UOB benchmark, ABC-Net achieved >95% accuracy, whereas the deep learning baseline (Img2mol) dropped to 78.2%, indicating better generalization.
- Data Efficiency: The model reached ~95% performance with only 80,000 training images, proving it requires orders of magnitude less data than captioning-based models (which often use millions).
- Noise Robustness: Performance remained stable (<2% drop) with noise levels up to $p=0.1$. Even at extreme noise ($p=0.6$), Tanimoto similarity remained high, suggesting the model recovers most substructures even when exact matches fail.
Reproducibility Details
Data
The authors constructed a synthetic dataset because labeled pixel-wise OCSR data is unavailable.
- Source: ChEMBL database
- Filtering: Excluded molecules with >50 non-H atoms or rare atom types/charges (<1000 occurrences).
- Sampling: 100,000 unique SMILES selected such that every atom type/charge appears in at least 1,000 compounds.
- Generation: Images generated via RDKit and Indigo libraries.
- Augmentation: Varied bond thickness, label mode, orientation, and aromaticity markers.
- Resolution: $512 \x512$ pixels.
- Noise: Salt-and-pepper noise added during training ($P$ = prob of background flip, $Q = 50P$).
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Training | ChEMBL (RDKit/Indigo) | 80k | 8:1:1 split (Train/Val/Test) |
| Evaluation | UOB Dataset | ~5.7k images | External benchmark from Univ. of Birmingham |
Algorithms
1. Keypoint Detection (Heatmaps)
- Down-sampling: Input $512 \x512$ → Output $102 \x102$ (stride 5).
- Label Softening: To handle discretization error, ground truth peaks are set to 1, first-order neighbors to 0.95, others to 0.
- Loss Function: Penalty-reduced pixel-wise binary focal loss (variants of CornerNet loss).
- $\alpha=2$ (focal parameter).
- Weight balancing: Classes <10% frequency weighted 10x.
2. Bond Direction Classification
- Angle Binning: $360°$ divided into 60 intervals.
- Inference: A bond is detected if the angle probability is a local maximum and exceeds a threshold.
- Non-Maximum Suppression (NMS): Required for opposite angles (e.g., $30°$ and $210°$) representing the same non-stereo bond.
3. Multi-Task Weighting
- Uses Kendall’s uncertainty weighting to balance 8 different loss terms (atom det, bond det, atom type, charge, H-count, bond angle, bond type, bond length).
Models
Architecture: ABC-Net (Custom U-Net / FCN)
- Input: $512 \x512 \x1$ (Grayscale).
- Contracting Path: 5 steps. Each step has conv-blocks + $2 \x2$ MaxPool.
- Expansive Path: 3 steps. Transpose-Conv upsampling + Concatenation (Skip Connections).
- Heads: Separate $1 \x1$ convs for each task map (Atom Heatmap, Bond Heatmap, Property Maps).
- Output Dimensions:
- Heatmaps: $(1, 102, 102)$
- Bond Angles: $(60, 102, 102)$
Evaluation
Metrics:
- Detection: Precision & Recall (Object detection level).
- Regression: Mean Absolute Error (MAE) for bond lengths.
- Structure Recovery:
- Accuracy: Exact SMILES match rate.
- Tanimoto: ECFP similarity (fingerprint overlap).
| Metric | ABC-Net | Img2mol (Baseline) | Notes |
|---|---|---|---|
| Accuracy (UOB) | 96.1% | 78.2% | Non-stereo subset |
| Accuracy (Indigo) | 96.4% | 89.5% | Non-stereo subset |
| Tanimoto (UOB) | 0.989 | 0.953 | Higher substructure recovery |
Hardware
- Training Configuration: 15 epochs, Batch size 64.
- Optimization: Adam Optimizer. LR $2.5 \x10^{-4}$ (first 5 epochs) → $2.5 \x10^{-5}$ (last 10).
- Compute: “High-Performance Computing Center” mentioned, specific GPU model not listed, but method described as “efficient” on GPU.
Citation
@article{zhangABCNetDivideandconquerBased2022,
title = {ABC-Net: A Divide-and-Conquer Based Deep Learning Architecture for {SMILES} Recognition from Molecular Images},
author = {Zhang, Xiao-Chen and Yi, Jia-Cai and Yang, Guo-Ping and Wu, Cheng-Kun and Hou, Ting-Jun and Cao, Dong-Sheng},
journal = {Briefings in Bioinformatics},
volume = {23},
number = {2},
pages = {bbac033},
year = {2022},
publisher = {Oxford University Press},
doi = {10.1093/bib/bbac033}
}