Paper Information
Citation: Dhaked, D. K., Ihlenfeldt, W.-D., Patel, H., Delannée, V., & Nicklaus, M. C. (2020). Toward a Comprehensive Treatment of Tautomerism in Chemoinformatics Including in InChI V2. Journal of Chemical Information and Modeling, 60(3), 1253-1275. https://doi.org/10.1021/acs.jcim.9b01080
Publication: Journal of Chemical Information and Modeling, 2020
Additional Resources:
- Tautomerizer Tool - Public web tool for testing tautomeric transformations
What kind of paper is this?
This is a Resource paper with strong Systematization elements. It provides a comprehensive catalog of 86 tautomeric transformation rules derived from experimental literature, designed to serve as a foundational resource for chemical database systems and the InChI V2 identifier standard. The systematic validation across 400+ million structures also makes it a benchmarking study for evaluating current chemoinformatics tools.
What is the motivation?
Chemical databases face a fundamental problem: the same molecule can appear multiple times under different identifiers simply because it exists in different tautomeric forms. For example, glucose’s ring-closed and open-chain forms are the same molecule, but current chemical identifiers (including InChI) often treat them as distinct compounds.
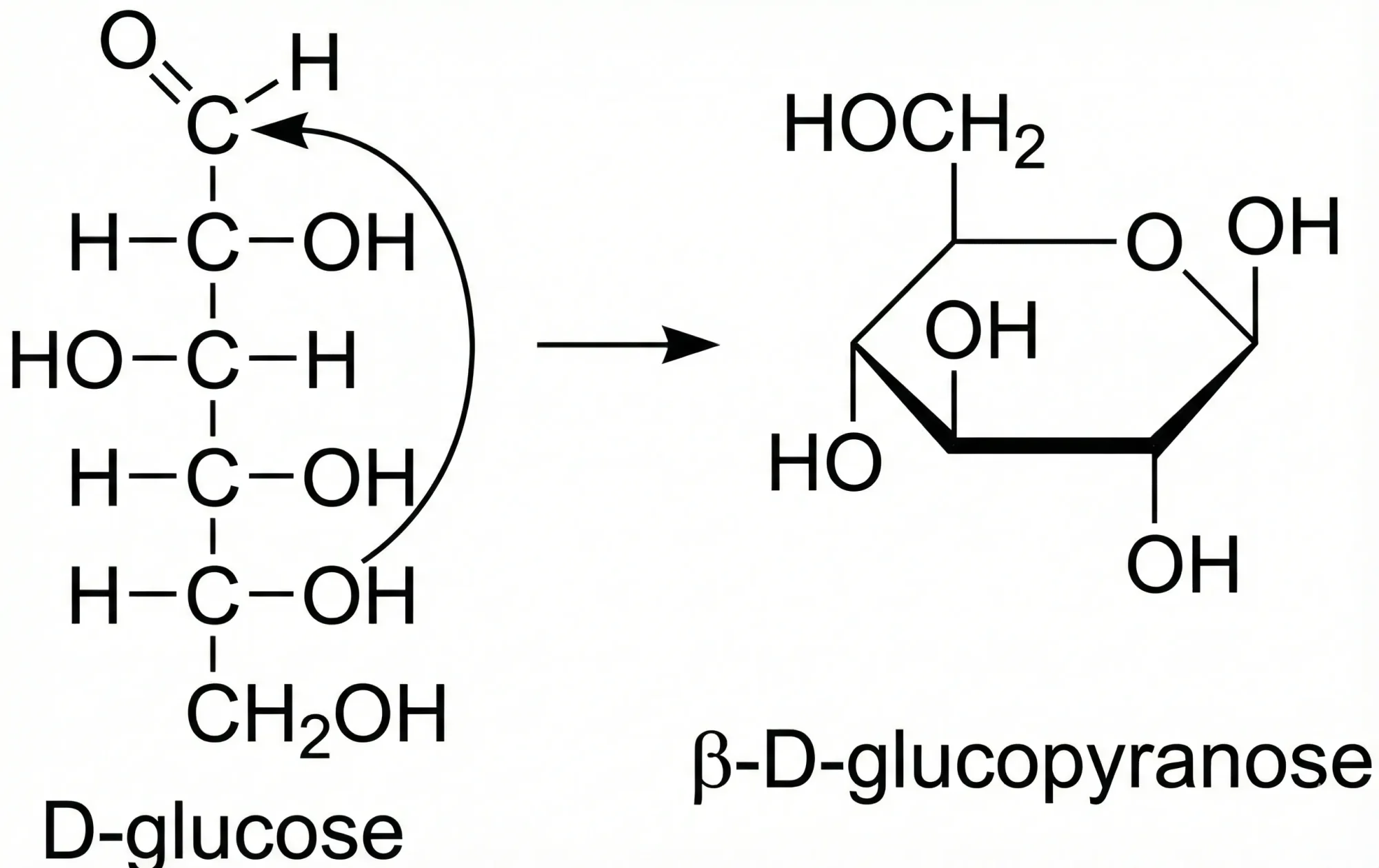
This creates three critical problems:
- Database redundancy: Millions of duplicate entries for the same chemical entities
- Search failures: Researchers miss relevant compounds during structure searches
- ML training issues: Machine learning models learn to treat tautomers as different molecules
The motivation for this work is to provide a comprehensive, experimentally-grounded rule set that enables InChI V2 to properly recognize tautomeric relationships, eliminating these problems at the identifier level.
What is the novelty here?
The key contributions are:
Comprehensive Rule Set: Development of 86 tautomeric transformation rules based on experimental literature, categorized into:
- 54 Prototropic rules (classic H-movement tautomerism)
- 21 Ring-Chain rules (cyclic/open-chain transformations)
- 11 Valence rules (structural rearrangements with valence changes)
Massive-Scale Validation: Testing these rules against nine major chemical databases totaling over 400 million structures to identify coverage gaps in current InChI implementations
Quantitative Assessment: Systematic measurement showing that current InChI (even with flexible settings) only achieves ~50% success in recognizing tautomeric relationships, with some new rules showing <2% success rates
Practical Tools: Creation of the Tautomerizer web tool for public use, demonstrating practical application of the rule set
The novelty lies in the systematic compilation and validation of transformation rules at a scale that reveals critical gaps in current chemical identification systems.
What experiments were performed?
Database Analysis
The researchers analyzed 9 chemical databases totaling 400+ million structures:
- Public databases: PubChem (largest), ChEMBL, DrugBank, PDB Ligands, SureChEMBL, AMS, ChemNavigator
- Private databases: CSD (Cambridge Structural Database), CSDB (NCI internal)
Methodology
Software: CACTVS Chemoinformatics Toolkit (versions 3.4.6.33 and 3.4.8.6)
Tautomer Generation Protocol:
- Algorithm: Single-step generation (apply transforms to input structure only, not recursive)
- Constraints: Max 10 tautomers per structure, 30-second CPU timeout per transform
- Format: All rules expressed as SMIRKS strings
- Stereochemistry: Stereocenters involved in tautomerism were flattened during transformation
Success Metrics:
- Complete InChI match: All tautomers share identical InChI
- Partial InChI match: At least two tautomers share an InChI
- Tested against three InChI configurations: Standard, Nonstandard (15T), Nonstandard (15T + KET)
Rule Coverage Analysis
For each of the 86 rules, the researchers:
- Applied the transformation to all molecules in each database
- Generated tautomers using the SMIRKS patterns
- Computed InChI identifiers for each tautomer
- Measured success rates (percentage of cases where InChI recognized the relationship)
Key Findings from Experiments
Rule Frequency: The most common rule PT_06_00 (1,3-heteroatom H-shift, covering keto-enol tautomerism) affects >70% of molecules across databases.
InChI Performance:
- Standard InChI: ~37% success rate
- Nonstandard InChI (15T): ~50% success rate
- Many newly defined rules: <2% success rate
Scale Impact: Implementing the full 86-rule set would triple the number of compounds recognized as having tautomeric relationships compared to current InChI.
What outcomes/conclusions?
Main Findings
Current Systems Are Inadequate: Even the most flexible InChI settings only achieve ~50% success in recognizing tautomeric relationships, with Standard InChI at ~37%
Massive Coverage Gap: The new rule set reveals millions of tautomeric relationships that current InChI completely misses, particularly for ring-chain and valence tautomerism
Implementation Requirement: InChI V2 will require a major redesign to handle the comprehensive rule set
Rule Validation: The 86-rule set, derived from experimental literature, provides a validated foundation for next-generation chemical identifiers
Implications
For Chemical Databases:
- Reduced redundancy through proper tautomer recognition
- Improved data quality and consistency
- More comprehensive structure search results
For Machine Learning:
- More accurate training data (tautomers properly grouped)
- Better molecular property prediction models
- Reduced dataset bias from tautomeric duplicates
For Chemoinformatics Tools:
- Blueprint for InChI V2 development
- Standardized rule set for tautomer generation
- Public tool (Tautomerizer) for practical use
Limitations Acknowledged
- Single-step generation only (doesn’t enumerate all possible tautomers recursively)
- 30-second timeout may miss complex transformations
- Some tautomeric preferences are context-dependent (pH, solvent) and can’t be captured by static rules alone
Future Directions
The paper lays groundwork for InChI V2 development, emphasizing that the comprehensive rule set necessitates algorithmic redesign.
Reproducibility Details
Data
Datasets Analyzed (400M+ total structures):
- PubChem: Largest public chemical database
- ChEMBL: Bioactive molecules with drug-like properties
- DrugBank: FDA-approved and experimental drugs
- PDB Ligands: Small molecules from protein structures
- SureChEMBL: Chemical structures from patents
- AMS: Screening samples
- ChemNavigator: Commercial chemical database
- CSD: Cambridge Structural Database (private)
- CSDB: NCI internal database (private)
Algorithms
Tautomer Generation:
- Method: Single-step SMIRKS-based transformations
- Constraints:
- Maximum 10 tautomers per input structure
- 30-second CPU timeout per transformation
- Stereochemistry flattening for affected centers
Rule Categories:
- Prototropic (PT): 54 rules for hydrogen movement
- Most common:
PT_06_00(1,3-heteroatom H-shift, >70% coverage)
- Most common:
- Ring-Chain (RC): 21 rules for cyclic/open-chain transformations
- Examples:
RC_03_00(pentose sugars),RC_04_01(hexose sugars)
- Examples:
- Valence (VT): 11 rules for valence changes
- Notable:
VT_02_00(tetrazole/azide, ~2.8M hits)
- Notable:
InChI Comparison:
- Standard InChI (default settings)
- Nonstandard InChI with
15Toption (mobile H) - Nonstandard InChI with
15TandKEToptions (keto-enol)
Evaluation
Success Metrics:
- Complete Match: All generated tautomers have identical InChI
- Partial Match: At least 2 tautomers share the same InChI
- Fail: All tautomers have different InChIs
Benchmark Results:
- Standard InChI: ~37% success rate across all rules
- Nonstandard (15T): ~50% success rate
- New rules: Many show <2% recognition by current InChI
Hardware
Software Environment:
- Toolkit: CACTVS Chemoinformatics Toolkit v3.4.6.33 and v3.4.8.6
- Hash Functions:
E_TAUTO_HASH(tautomer-invariant identifier)E_ISOTOPE_STEREO_HASH128(tautomer-sensitive identifier)
Note: The paper doesn’t specify computational hardware, as the analysis was performed using existing chemical databases.
