Key Contribution#
MARCEL provides the first comprehensive benchmark for conformer ensemble learning, demonstrating that explicitly modeling full conformer distributions significantly improves property prediction across drug-like molecules and organometallic catalysts.
Overview#
The Molecular Representation and Conformer Ensemble Learning (MARCEL) dataset provides 722K+ conformations across 76K+ molecules spanning four diverse chemical domains: drug-like molecules (Drugs-75K), organophosphorus ligands (Kraken), chiral catalysts (EE), and organometallic complexes (BDE). Unlike prior datasets that focus solely on drug-like molecules, MARCEL enables evaluation of conformer ensemble methods across both pharmaceutical and catalysis applications.
Strengths#
- Domain diversity: Beyond drug-like molecules, includes organometallics and catalysts rarely covered in existing benchmarks
- Ensemble-based: Provides full conformer ensembles with statistical weights
- DFT-quality energies: Drugs-75K features DFT-level conformers and energies (higher accuracy than GEOM-Drugs)
- Realistic scenarios: BDE subset models the practical constraint of lacking DFT-computed conformers for large catalyst systems
- Comprehensive baselines: Benchmarks 18 models across 1D (SMILES), 2D (graph), 3D (single conformer), and ensemble methods
- Property diversity: Covers ionization potential, electron affinity, electronegativity, ligand descriptors, and catalytic properties
Limitations#
- Regression only: All tasks are regression; no classification benchmarks
- Chemical space coverage: 76K molecules cannot represent full drug-like or catalyst chemical space
- Compute requirements: Working with large conformer ensembles demands significant computational resources
- Proprietary data: EE subset not publicly available (as of December 2025)
- DFT bottleneck: BDE demonstrates the practical limitation - single DFT optimization can take 2-3 days, making conformer-level DFT infeasible for large organometallics
Technical Notes#
Data Generation Pipeline#
Drugs-75K#
Source: GEOM-Drugs subset
Filtering:
- Minimum 5 rotatable bonds (focus on flexible molecules)
- Allowed elements: H, C, N, O, F, Si, P, S, Cl
Conformer generation:
- DFT-level calculations for both conformers and energies
- Higher accuracy than original GEOM-Drugs (semi-empirical GFN2-xTB)
Properties: Ionization Potential (IP), Electron Affinity (EA), Electronegativity (χ)
Kraken#
Source: Original Kraken dataset (1,552 monodentate organophosphorus(III) ligands)
Properties: 5 of 78 available properties
- $B_5$: Ligand sterics (buried volume)
- $L$: Ligand electronics
- $\text{Bur}B_5$: Buried volume variant
- $\text{Bur}L$: Electronic parameter variant
EE (Enantiomeric Excess)#
Generation method: Q2MM (Quantum-guided Molecular Mechanics)
Molecules: 872 Rhodium (Rh)-bound atropisomeric catalysts from chiral bisphosphine
Property: Enantiomeric excess (EE) for asymmetric catalysis
Availability: Proprietary-only (not publicly available as of December 2025)
BDE (Bond Dissociation Energy)#
Molecules: 5,195 organometallic catalysts (ML₁L₂ structure)
Initial conformers: OpenBabel with geometric optimization
Energies: DFT calculations
Property: Electronic dissociation energy (difference between bound and unbound states)
Key constraint: DFT optimization for full conformer ensembles computationally infeasible (2-3 days per molecule)
Benchmark Setup#
Task: Predict molecular properties from structure using different representation strategies (1D/2D/3D/Ensemble)
Data splits: Not explicitly specified in the available information; standard train/validation/test splits used
Hyperparameters: Tuned per model (specific optimization method not detailed in available documentation)
Model categories:
- 1D Models: SMILES-based (Random Forest on Morgan fingerprints, LSTM, Transformer)
- 2D Models: Graph-based (GIN, GIN+VN, ChemProp, GraphGPS)
- 3D Models: Single conformer (SchNet, DimeNet++, GemNet, PaiNN, ClofNet, LEFTNet)
- Ensemble Models: Full conformer ensemble (same 3D architectures, aggregating over all conformers)
Evaluation metric: Mean Absolute Error (MAE) for all tasks
Key Findings#
Ensemble superiority: Across all three benchmarks, ensemble methods (processing full conformer distributions) consistently outperform single-conformer 3D models, with the largest improvements on:
- Drugs-75K: Ensemble GemNet achieves 🥇 0.4066 eV (IP) vs 🥈 0.4069 eV (single conformer)
- Kraken: Ensemble PaiNN achieves 🥇 0.2225 (B₅) vs 0.3443 (single conformer)
- EE: Ensemble GemNet achieves 🥇 11.61% vs 18.03% (single conformer)
3D vs 2D: 3D models generally outperform 2D graph models, especially for conformationally-sensitive properties
Model architecture: GemNet and PaiNN architectures consistently top-ranked across tasks
Benchmarks#
Ionization Potential (Drugs-75K)#
Predict ionization potential from molecular structure
Subset: Drugs-75K
| Rank | Model | MAE (eV) |
|---|
| 🥇 1 | Ensemble - GemNet
GemNet on full conformer ensemble | 0.4066 |
| 🥈 2 | 3D - GemNet
Geometry-enhanced message passing (single conformer) | 0.4069 |
| 🥉 3 | Ensemble - DimeNet++
DimeNet++ on full conformer ensemble | 0.4126 |
| 4 | Ensemble - LEFTNet
LEFTNet on full conformer ensemble | 0.4149 |
| 5 | 3D - LEFTNet
Local Environment Feature Transformer (single conformer) | 0.4174 |
| 6 | Ensemble - ClofNet
ClofNet on full conformer ensemble | 0.428 |
| 7 | 2D - GraphGPS
Graph Transformer with positional encodings | 0.4351 |
| 8 | 2D - GIN
Graph Isomorphism Network | 0.4354 |
| 9 | 2D - GIN+VN
GIN with Virtual Nodes | 0.4361 |
| 10 | 3D - ClofNet
Conformation-ensemble learning network (single conformer) | 0.4393 |
| 11 | 3D - SchNet
Continuous-filter convolutional network (single conformer) | 0.4394 |
| 12 | 3D - DimeNet++
Directional message passing network (single conformer) | 0.4441 |
| 13 | Ensemble - SchNet
SchNet on full conformer ensemble | 0.4452 |
| 14 | Ensemble - PaiNN
PaiNN on full conformer ensemble | 0.4466 |
| 15 | 3D - PaiNN
Polarizable Atom Interaction Network (single conformer) | 0.4505 |
| 16 | 2D - ChemProp
Message Passing Neural Network | 0.4595 |
| 17 | 1D - LSTM
LSTM on SMILES sequences | 0.4788 |
| 18 | 1D - Random forest
Random Forest on Morgan fingerprints | 0.4987 |
| 19 | 1D - Transformer
Transformer on SMILES sequences | 0.6617 |
Electron Affinity (Drugs-75K)#
Predict electron affinity from molecular structure
Subset: Drugs-75K
| Rank | Model | MAE (eV) |
|---|
| 🥇 1 | Ensemble - GemNet
GemNet on full conformer ensemble | 0.391 |
| 🥈 2 | 3D - GemNet
Geometry-enhanced message passing (single conformer) | 0.3922 |
| 🥉 3 | Ensemble - DimeNet++
DimeNet++ on full conformer ensemble | 0.3944 |
| 4 | Ensemble - LEFTNet
LEFTNet on full conformer ensemble | 0.3953 |
| 5 | 3D - LEFTNet
Local Environment Feature Transformer (single conformer) | 0.3964 |
| 6 | Ensemble - ClofNet
ClofNet on full conformer ensemble | 0.4033 |
| 7 | 2D - GraphGPS
Graph Transformer with positional encodings | 0.4085 |
| 8 | 2D - GIN
Graph Isomorphism Network | 0.4169 |
| 9 | 2D - GIN+VN
GIN with Virtual Nodes | 0.4169 |
| 10 | 3D - SchNet
Continuous-filter convolutional network (single conformer) | 0.4207 |
| 11 | 3D - DimeNet++
Directional message passing network (single conformer) | 0.4233 |
| 12 | Ensemble - SchNet
SchNet on full conformer ensemble | 0.4232 |
| 13 | 3D - ClofNet
Conformation-ensemble learning network (single conformer) | 0.4251 |
| 14 | Ensemble - PaiNN
PaiNN on full conformer ensemble | 0.4269 |
| 15 | 2D - ChemProp
Message Passing Neural Network | 0.4417 |
| 16 | 3D - PaiNN
Polarizable Atom Interaction Network (single conformer) | 0.4495 |
| 17 | 1D - LSTM
LSTM on SMILES sequences | 0.4648 |
| 18 | 1D - Random forest
Random Forest on Morgan fingerprints | 0.4747 |
| 19 | 1D - Transformer
Transformer on SMILES sequences | 0.585 |
Electronegativity (Drugs-75K)#
Predict electronegativity (χ) from molecular structure
Subset: Drugs-75K
| Rank | Model | MAE (eV) |
|---|
| 🥇 1 | 3D - GemNet
Geometry-enhanced message passing (single conformer) | 0.197 |
| 🥈 2 | Ensemble - GemNet
GemNet on full conformer ensemble | 0.2027 |
| 🥉 3 | Ensemble - LEFTNet
LEFTNet on full conformer ensemble | 0.2069 |
| 4 | 3D - LEFTNet
Local Environment Feature Transformer (single conformer) | 0.2083 |
| 5 | Ensemble - ClofNet
ClofNet on full conformer ensemble | 0.2199 |
| 6 | 2D - GraphGPS
Graph Transformer with positional encodings | 0.2212 |
| 7 | 3D - SchNet
Continuous-filter convolutional network (single conformer) | 0.2243 |
| 8 | Ensemble - SchNet
SchNet on full conformer ensemble | 0.2243 |
| 9 | 2D - GIN
Graph Isomorphism Network | 0.226 |
| 10 | 2D - GIN+VN
GIN with Virtual Nodes | 0.2267 |
| 11 | Ensemble - DimeNet++
DimeNet++ on full conformer ensemble | 0.2267 |
| 12 | Ensemble - PaiNN
PaiNN on full conformer ensemble | 0.2294 |
| 13 | 3D - PaiNN
Polarizable Atom Interaction Network (single conformer) | 0.2324 |
| 14 | 3D - ClofNet
Conformation-ensemble learning network (single conformer) | 0.2378 |
| 15 | 3D - DimeNet++
Directional message passing network (single conformer) | 0.2436 |
| 16 | 2D - ChemProp
Message Passing Neural Network | 0.2441 |
| 17 | 1D - LSTM
LSTM on SMILES sequences | 0.2505 |
| 18 | 1D - Random forest
Random Forest on Morgan fingerprints | 0.2732 |
| 19 | 1D - Transformer
Transformer on SMILES sequences | 0.4073 |
B₅ Sterimol Parameter (Kraken)#
Predict B₅ sterimol descriptor for organophosphorus ligands
Subset: Kraken
| Rank | Model | MAE |
|---|
| 🥇 1 | Ensemble - PaiNN
PaiNN on full conformer ensemble | 0.2225 |
| 🥈 2 | Ensemble - GemNet
GemNet on full conformer ensemble | 0.2313 |
| 🥉 3 | Ensemble - DimeNet++
DimeNet++ on full conformer ensemble | 0.263 |
| 4 | Ensemble - LEFTNet
LEFTNet on full conformer ensemble | 0.2644 |
| 5 | Ensemble - SchNet
SchNet on full conformer ensemble | 0.2704 |
| 6 | 3D - GemNet
Geometry-enhanced message passing (single conformer) | 0.2789 |
| 7 | 3D - LEFTNet
Local Environment Feature Transformer (single conformer) | 0.3072 |
| 8 | 2D - GIN
Graph Isomorphism Network | 0.3128 |
| 9 | Ensemble - ClofNet
ClofNet on full conformer ensemble | 0.3228 |
| 10 | 3D - SchNet
Continuous-filter convolutional network (single conformer) | 0.3293 |
| 11 | 3D - PaiNN
Polarizable Atom Interaction Network (single conformer) | 0.3443 |
| 12 | 2D - GraphGPS
Graph Transformer with positional encodings | 0.345 |
| 13 | 3D - DimeNet++
Directional message passing network (single conformer) | 0.351 |
| 14 | 2D - GIN+VN
GIN with Virtual Nodes | 0.3567 |
| 15 | 1D - Random forest
Random Forest on Morgan fingerprints | 0.476 |
| 16 | 2D - ChemProp
Message Passing Neural Network | 0.485 |
| 17 | 3D - ClofNet
Conformation-ensemble learning network (single conformer) | 0.4873 |
| 18 | 1D - LSTM
LSTM on SMILES sequences | 0.4879 |
| 19 | 1D - Transformer
Transformer on SMILES sequences | 0.9611 |
L Sterimol Parameter (Kraken)#
Predict L sterimol descriptor for organophosphorus ligands
Subset: Kraken
| Rank | Model | MAE |
|---|
| 🥇 1 | Ensemble - GemNet
GemNet on full conformer ensemble | 0.3386 |
| 🥈 2 | Ensemble - DimeNet++
DimeNet++ on full conformer ensemble | 0.3468 |
| 🥉 3 | Ensemble - PaiNN
PaiNN on full conformer ensemble | 0.3619 |
| 4 | Ensemble - LEFTNet
LEFTNet on full conformer ensemble | 0.3643 |
| 5 | 3D - GemNet
Geometry-enhanced message passing (single conformer) | 0.3754 |
| 6 | 2D - GIN
Graph Isomorphism Network | 0.4003 |
| 7 | 3D - DimeNet++
Directional message passing network (single conformer) | 0.4174 |
| 8 | 1D - Random forest
Random Forest on Morgan fingerprints | 0.4303 |
| 9 | Ensemble - SchNet
SchNet on full conformer ensemble | 0.4322 |
| 10 | 2D - GIN+VN
GIN with Virtual Nodes | 0.4344 |
| 11 | 2D - GraphGPS
Graph Transformer with positional encodings | 0.4363 |
| 12 | 3D - PaiNN
Polarizable Atom Interaction Network (single conformer) | 0.4471 |
| 13 | Ensemble - ClofNet
ClofNet on full conformer ensemble | 0.4485 |
| 14 | 3D - LEFTNet
Local Environment Feature Transformer (single conformer) | 0.4493 |
| 15 | 1D - LSTM
LSTM on SMILES sequences | 0.5142 |
| 16 | 2D - ChemProp
Message Passing Neural Network | 0.5452 |
| 17 | 3D - SchNet
Continuous-filter convolutional network (single conformer) | 0.5458 |
| 18 | 3D - ClofNet
Conformation-ensemble learning network (single conformer) | 0.6417 |
| 19 | 1D - Transformer
Transformer on SMILES sequences | 0.8389 |
Buried B₅ Parameter (Kraken)#
Predict buried B₅ sterimol descriptor for organophosphorus ligands
Subset: Kraken
| Rank | Model | MAE |
|---|
| 🥇 1 | Ensemble - GemNet
GemNet on full conformer ensemble | 0.1589 |
| 🥈 2 | Ensemble - PaiNN
PaiNN on full conformer ensemble | 0.1693 |
| 🥉 3 | 2D - GIN
Graph Isomorphism Network | 0.1719 |
| 4 | 3D - GemNet
Geometry-enhanced message passing (single conformer) | 0.1782 |
| 5 | Ensemble - DimeNet++
DimeNet++ on full conformer ensemble | 0.1783 |
| 6 | Ensemble - SchNet
SchNet on full conformer ensemble | 0.2024 |
| 7 | Ensemble - LEFTNet
LEFTNet on full conformer ensemble | 0.2017 |
| 8 | 2D - GraphGPS
Graph Transformer with positional encodings | 0.2066 |
| 9 | 3D - DimeNet++
Directional message passing network (single conformer) | 0.2097 |
| 10 | Ensemble - ClofNet
ClofNet on full conformer ensemble | 0.2178 |
| 11 | 3D - LEFTNet
Local Environment Feature Transformer (single conformer) | 0.2176 |
| 12 | 3D - SchNet
Continuous-filter convolutional network (single conformer) | 0.2295 |
| 13 | 3D - PaiNN
Polarizable Atom Interaction Network (single conformer) | 0.2395 |
| 14 | 2D - GIN+VN
GIN with Virtual Nodes | 0.2422 |
| 15 | 1D - Random forest
Random Forest on Morgan fingerprints | 0.2758 |
| 16 | 1D - LSTM
LSTM on SMILES sequences | 0.2813 |
| 17 | 3D - ClofNet
Conformation-ensemble learning network (single conformer) | 0.2884 |
| 18 | 2D - ChemProp
Message Passing Neural Network | 0.3002 |
| 19 | 1D - Transformer
Transformer on SMILES sequences | 0.4929 |
Buried L Parameter (Kraken)#
Predict buried L sterimol descriptor for organophosphorus ligands
Subset: Kraken
| Rank | Model | MAE |
|---|
| 🥇 1 | Ensemble - GemNet
GemNet on full conformer ensemble | 0.0947 |
| 🥈 2 | Ensemble - DimeNet++
DimeNet++ on full conformer ensemble | 0.1185 |
| 🥉 3 | 2D - GIN
Graph Isomorphism Network | 0.12 |
| 4 | Ensemble - PaiNN
PaiNN on full conformer ensemble | 0.1324 |
| 5 | Ensemble - LEFTNet
LEFTNet on full conformer ensemble | 0.1386 |
| 6 | Ensemble - SchNet
SchNet on full conformer ensemble | 0.1443 |
| 7 | 3D - LEFTNet
Local Environment Feature Transformer (single conformer) | 0.1486 |
| 8 | 2D - GraphGPS
Graph Transformer with positional encodings | 0.15 |
| 9 | 1D - Random forest
Random Forest on Morgan fingerprints | 0.1521 |
| 10 | 3D - DimeNet++
Directional message passing network (single conformer) | 0.1526 |
| 11 | Ensemble - ClofNet
ClofNet on full conformer ensemble | 0.1548 |
| 12 | 3D - GemNet
Geometry-enhanced message passing (single conformer) | 0.1635 |
| 13 | 3D - PaiNN
Polarizable Atom Interaction Network (single conformer) | 0.1673 |
| 14 | 2D - GIN+VN
GIN with Virtual Nodes | 0.1741 |
| 15 | 3D - SchNet
Continuous-filter convolutional network (single conformer) | 0.1861 |
| 16 | 1D - LSTM
LSTM on SMILES sequences | 0.1924 |
| 17 | 2D - ChemProp
Message Passing Neural Network | 0.1948 |
| 18 | 3D - ClofNet
Conformation-ensemble learning network (single conformer) | 0.2529 |
| 19 | 1D - Transformer
Transformer on SMILES sequences | 0.2781 |
Enantioselectivity (EE)#
Predict enantiomeric excess for Rh-catalyzed asymmetric reactions
Subset: EE
| Rank | Model | MAE (%) |
|---|
| 🥇 1 | Ensemble - GemNet
GemNet on full conformer ensemble | 11.61 |
| 🥈 2 | Ensemble - DimeNet++
DimeNet++ on full conformer ensemble | 12.03 |
| 🥉 3 | Ensemble - PaiNN
PaiNN on full conformer ensemble | 13.56 |
| 4 | Ensemble - ClofNet
ClofNet on full conformer ensemble | 13.96 |
| 5 | Ensemble - SchNet
SchNet on full conformer ensemble | 14.22 |
| 6 | 3D - DimeNet++
Directional message passing network (single conformer) | 14.64 |
| 7 | 3D - SchNet
Continuous-filter convolutional network (single conformer) | 17.74 |
| 8 | 3D - GemNet
Geometry-enhanced message passing (single conformer) | 18.03 |
| 9 | Ensemble - LEFTNet
LEFTNet on full conformer ensemble | 18.42 |
| 10 | 3D - LEFTNet
Local Environment Feature Transformer (single conformer) | 19.8 |
| 11 | 3D - PaiNN
Polarizable Atom Interaction Network (single conformer) | 20.24 |
| 12 | 3D - ClofNet
Conformation-ensemble learning network (single conformer) | 33.95 |
| 13 | 2D - ChemProp
Message Passing Neural Network | 61.03 |
| 14 | 1D - Random forest
Random Forest on Morgan fingerprints | 61.3 |
| 15 | 2D - GraphGPS
Graph Transformer with positional encodings | 61.63 |
| 16 | 1D - Transformer
Transformer on SMILES sequences | 62.08 |
| 17 | 2D - GIN
Graph Isomorphism Network | 62.31 |
| 18 | 2D - GIN+VN
GIN with Virtual Nodes | 62.38 |
| 19 | 1D - LSTM
LSTM on SMILES sequences | 64.01 |
Bond Dissociation Energy (BDE)#
Predict metal-ligand bond dissociation energy for organometallic catalysts
Subset: BDE
| Rank | Model | MAE (kcal/mol) |
|---|
| 🥇 1 | 3D - DimeNet++
Directional message passing network (single conformer) | 1.45 |
| 🥈 2 | Ensemble - DimeNet++
DimeNet++ on full conformer ensemble | 1.47 |
| 🥉 3 | 3D - LEFTNet
Local Environment Feature Transformer (single conformer) | 1.53 |
| 4 | Ensemble - LEFTNet
LEFTNet on full conformer ensemble | 1.53 |
| 5 | Ensemble - GemNet
GemNet on full conformer ensemble | 1.61 |
| 6 | 3D - GemNet
Geometry-enhanced message passing (single conformer) | 1.65 |
| 7 | Ensemble - PaiNN
PaiNN on full conformer ensemble | 1.87 |
| 8 | Ensemble - SchNet
SchNet on full conformer ensemble | 1.97 |
| 9 | Ensemble - ClofNet
ClofNet on full conformer ensemble | 2.01 |
| 10 | 3D - PaiNN
Polarizable Atom Interaction Network (single conformer) | 2.13 |
| 11 | 2D - GraphGPS
Graph Transformer with positional encodings | 2.48 |
| 12 | 3D - SchNet
Continuous-filter convolutional network (single conformer) | 2.55 |
| 13 | 3D - ClofNet
Conformation-ensemble learning network (single conformer) | 2.61 |
| 14 | 2D - GIN
Graph Isomorphism Network | 2.64 |
| 15 | 2D - ChemProp
Message Passing Neural Network | 2.66 |
| 16 | 2D - GIN+VN
GIN with Virtual Nodes | 2.74 |
| 17 | 1D - LSTM
LSTM on SMILES sequences | 2.83 |
| 18 | 1D - Random forest
Random Forest on Morgan fingerprints | 3.03 |
| 19 | 1D - Transformer
Transformer on SMILES sequences | 10.08 |
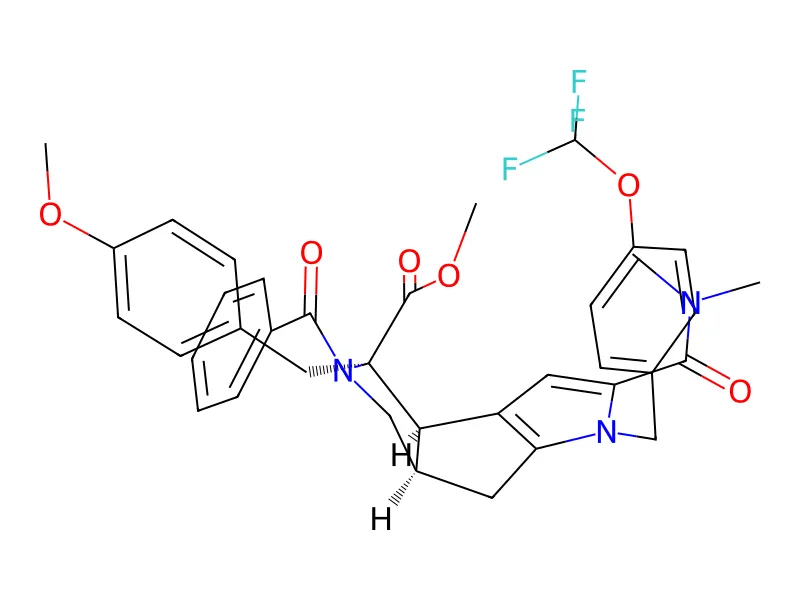
![Example conformer from Drugs-75K (SMILES: `COC(=O)[C@@]1(Cc2ccc(OC)cc2)[C@H]2c3cc(C(=O)N(C)C)n(Cc4ccc(OC(F)(F)F)cc4)c3C[C@H]2CN1C(=O)c1ccccc1`; IUPAC: methyl (2R,3R,6R)-4-benzoyl-10-(dimethylcarbamoyl)-3-[(4-methoxyphenyl)methyl]-9-[[4-(trifluoromethoxy)phenyl]methyl]-4,9-diazatricyclo[6.3.0.02,6]undeca-1(8),10-diene-3-carboxylate)](/img/marcel-drugs-75k-example-conformer.webp)