Paper Information
Citation: Krasnov, L., Khokhlov, I., Fedorov, M. V., & Sosnin, S. (2021). Transformer-based artificial neural networks for the conversion between chemical notations. Scientific Reports, 11(1), 14798. https://doi.org/10.1038/s41598-021-94082-y
Publication: Scientific Reports 2021
Additional Resources:
What kind of paper is this?
This is primarily a Method paper with significant elements of Position.
- Method: The authors propose a specific neural architecture (Transformer with custom tokenization) and a verification pipeline (round-trip check) to solve the SMILES $\leftrightarrow$ IUPAC translation task. They rigorously benchmark this against rule-based baselines (OPSIN).
- Position: The authors explicitly argue for a paradigm shift, suggesting that “heavy” neural architectures should replace complex, costly rule-based legacy systems even for “exact” algorithmic tasks.
What is the motivation?
- Complexity of Naming: Generating IUPAC names manually is error-prone and requires deep algorithmic knowledge.
- Lack of Open Source Tools: While open-source tools exist for Name-to-Structure (e.g., OPSIN), there were no open-source tools for the inverse “Structure-to-Name” conversion at the time of writing.
- Cost of Development: Developing rule-based converters “from scratch” is prohibitively expensive and time-consuming compared to training a neural model on existing data.
What is the novelty here?
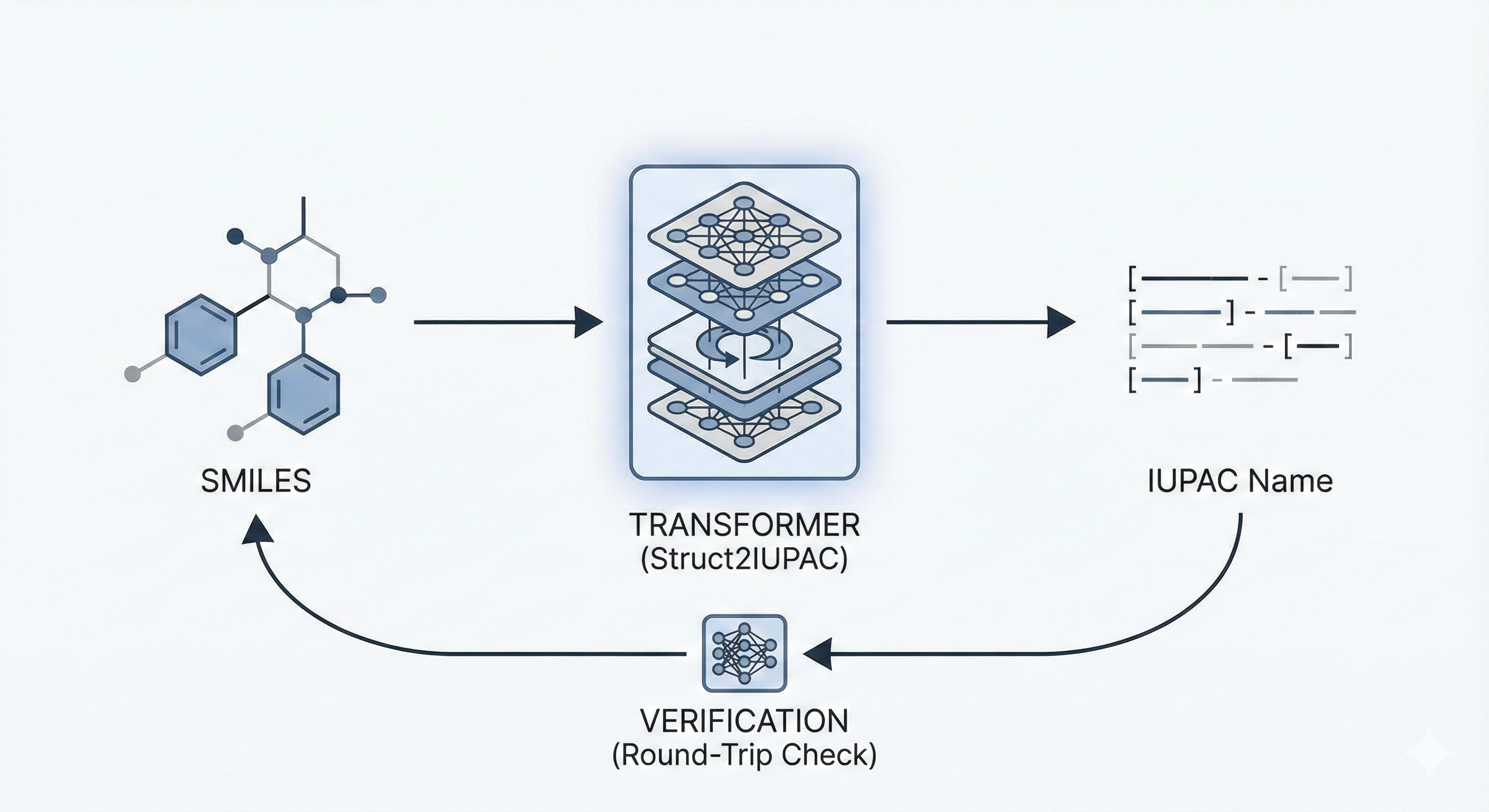
- Struct2IUPAC: The first effective open-source neural model for converting SMILES to IUPAC names, treating chemical translation as a Neural Machine Translation (NMT) problem.
- Verification Loop: A novel inference pipeline that generates multiple candidates via beam search and validates them using a reverse converter (OPSIN) to ensure the generated name maps back to the original structure.
- Custom Tokenization: A manually curated rule-based tokenizer for IUPAC names that handles specific chemical suffixes, prefixes, and stereochemical markers.
What experiments were performed?
- Accuracy Benchmarking: The models were tested on a held-out subset of 100,000 molecules from PubChem. The authors measured accuracy across different beam sizes (1, 3, 5).
- Comparison to Rules: The neural IUPAC2Struct model was compared directly against the rule-based OPSIN tool.
- Stress Testing:
- Sequence Length: Evaluated performance across varying token lengths, identifying a “sweet spot” (10-60 tokens) and failure modes for very short (e.g., methane) or long molecules.
- Stereochemistry: Tested on “stereo-dense” compounds, observing a performance drop but generally robust handling of stereocenters.
- Tautomers: Verified the model’s ability to handle different tautomeric forms (e.g., Guanine and Uracil variants).
- Latency Analysis: Benchmarked inference speeds on CPU vs. GPU relative to output sequence length.
What were the outcomes and conclusions drawn?
- High Accuracy: The Struct2IUPAC model achieved 98.9% accuracy (Beam 5 with verification). The reverse model (IUPAC2Struct) achieved 99.1%, comparable to OPSIN’s 99.4%.
- Chemical “Intuition”: The model demonstrated an ability to infer logic, correctly generating multiple valid IUPAC names for single molecules where naming ambiguity exists (e.g., parent group selection).
- Production Readiness: Inference on GPU takes less than 0.5 seconds even for long names, making it viable for production use.
- Paradigm Shift: The authors conclude that neural networks are a viable, cost-effective alternative to developing rule-based algorithms for legacy notation conversion.
Reproducibility Details
Data
The study utilized the PubChem database.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Total | PubChem | ~95M | Filtered for RDKit compatibility |
| Training | Split A | 47,312,235 | Random 50% split |
| Testing | Split B | 47,413,850 | Random 50% split |
- Cleaning: Molecules that could not be processed by RDKit were removed. Molecules containing tokens not in the tokenizer (e.g., aromatic selenium) were excluded.
- Availability: A subset of 100,000 test molecules is available on Zenodo and GitHub.
Algorithms
- Tokenization:
- SMILES: Character-based tokenization.
- IUPAC: Custom rule-based tokenizer splitting suffixes (
-one,-al), prefixes (-oxy,-di), and special symbols ((,),R(S)).
- Verification Step:
- Generate $N$ names using Beam Search ($N=5$).
- Reverse translate the candidate name using OPSIN.
- Check if the OPSIN structure matches the original input SMILES.
- Display the first verified match; otherwise, report failure.
Models
- Architecture: Standard Transformer with 6 encoder layers and 6 decoder layers.
- Hyperparameters:
- Attention Heads: 8
- Attention Dimension ($d_{\text{model}}$): 512
- Feed-Forward Dimension ($d_{\text{ff}}$): 2048
- Training: Two separate models were trained:
Struct2IUPAC(SMILES $\to$ IUPAC) andIUPAC2Struct(IUPAC $\to$ SMILES).
Evaluation
Evaluation was performed on a random subset of 100,000 molecules from the test set.
| Metric | Task | Beam Size | Accuracy |
|---|---|---|---|
| Exact Match | Struct2IUPAC | 1 | 96.1% |
| Exact Match | Struct2IUPAC | 5 | 98.9% |
| Exact Match | IUPAC2Struct | 1 | 96.6% |
| Exact Match | IUPAC2Struct | 5 | 99.1% |
- Robustness: Accuracy drops significantly for augmented (non-canonical) SMILES (37.16%) and stereo-enriched compounds (66.52%).
Hardware
- Training Infrastructure: 4 $\times$ Tesla V100 GPUs and 36 CPUs.
- Training Time: Approximately 10 days under full load.
- Inference Speed: <0.5s per molecule on GPU; scale is linear with output token length.
Citation
@article{krasnovTransformerbasedArtificialNeural2021a,
title = {Transformer-Based Artificial Neural Networks for the Conversion between Chemical Notations},
author = {Krasnov, Lev and Khokhlov, Ivan and Fedorov, Maxim V. and Sosnin, Sergey},
year = 2021,
month = jul,
journal = {Scientific Reports},
volume = {11},
number = {1},
pages = {14798},
publisher = {Nature Publishing Group},
doi = {10.1038/s41598-021-94082-y}
}