What kind of paper is this?
This is primarily a Methodological paper, as it proposes a specific neural architecture (GP-MoLFormer) and a novel fine-tuning algorithm (Pair-tuning) for molecular generation. It validates these contributions against standard baselines (e.g., JT-VAE, MolGen-7b).
It also contains a secondary Theoretical contribution by establishing an empirical scaling law that relates inference compute (generation size) to the novelty of the generated molecules.
What is the motivation?
While large language models (LLMs) have transformed text generation, the impact of training data scale and memorization on molecular generative models remains under-explored. Specifically, there is a need to understand how training on billion-scale datasets affects the novelty of generated molecules and whether biases in public databases (like ZINC and PubChem) perpetuate memorization. Furthermore, existing optimization methods often require computationally expensive property predictors or reinforcement learning loops; there is a practical need for more efficient “prompt-based” optimization techniques.
What is the novelty here?
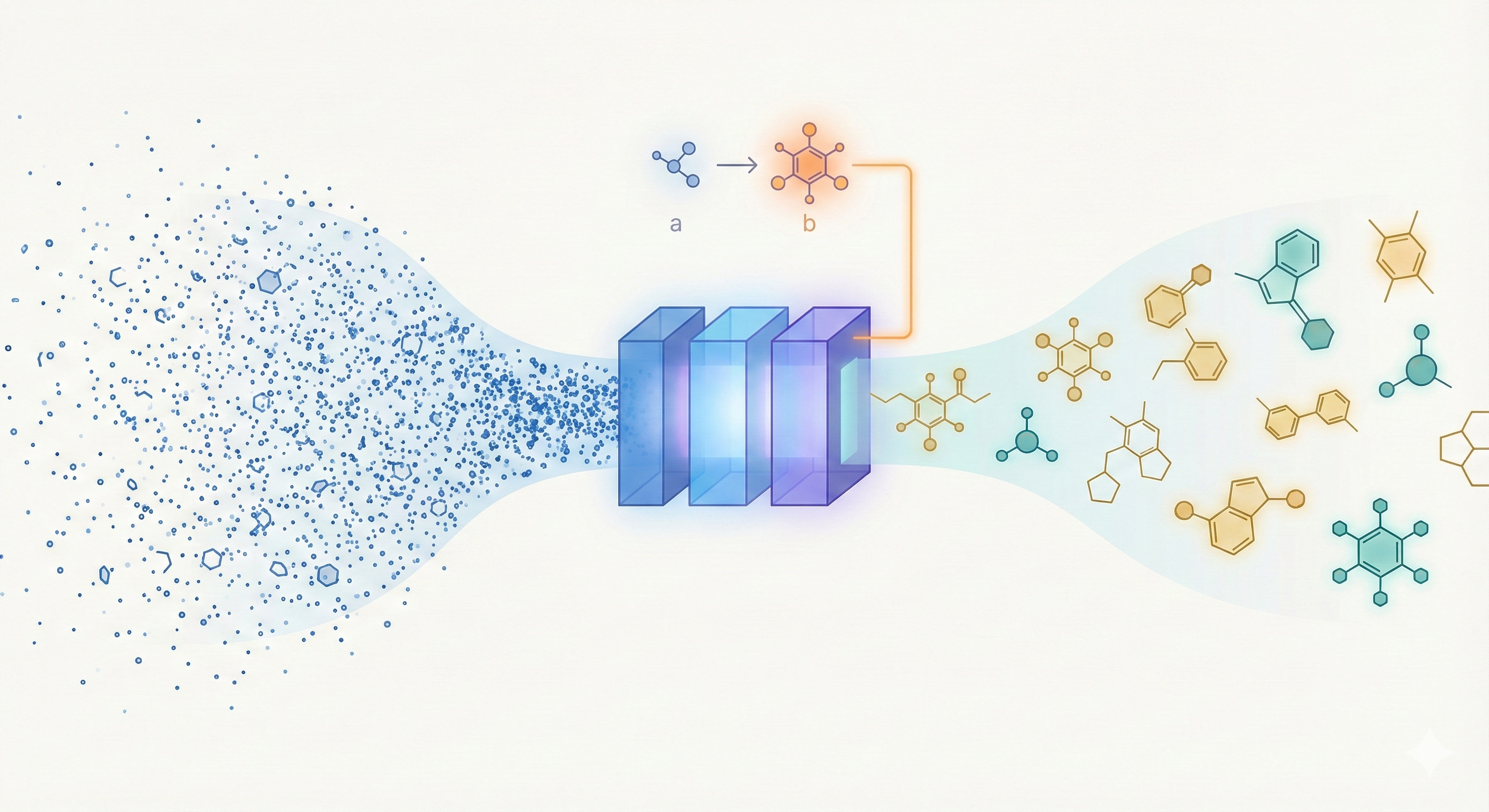
- Architecture: The application of a linear-attention transformer decoder with Rotary Positional Embeddings (RoPE) to generative chemistry, allowing for efficient training on 1.1 billion SMILES.
- Pair-Tuning: A novel, parameter-efficient fine-tuning method that uses property-ordered molecular pairs to learn “soft prompts” for optimization without updating the base model weights.
- Scaling Analysis: The first extensive investigation into the trade-off between inference compute (up to 10B generations) and chemical novelty, establishing that novelty follows an exponential decay law as generation size increases.
What experiments were performed?
The authors evaluated GP-MoLFormer on three distinct tasks:
- De Novo Generation: Comparing validity, uniqueness, and novelty against baselines (CharRNN, VAE, LIMO, MolGen-7b) on a held-out test set.
- Scaffold-Constrained Decoration: Generating molecules from DRD2 active binder scaffolds and measuring the hit rate of active compounds.
- Property-Guided Optimization: Using Pair-tuning to optimize for Drug-likeness (QED), Penalized logP, and DRD2 binding activity.
Additionally, they performed a Scaling Study:
- Comparing models trained on raw (1.1B) vs. de-duplicated (650M) data.
- Generating up to 10 billion molecules to fit empirical scaling laws for novelty.
What outcomes/conclusions?
- Performance: GP-MoLFormer achieves state-of-the-art or comparable performance on generation metrics, showing high internal diversity and validity (>99%) even at massive scales.
- Pair-Tuning Efficacy: The proposed pair-tuning method effectively optimizes properties (e.g., improving DRD2 activity scores significantly over baselines like Gargoyles) without requiring full model fine-tuning or external reward loops.
- Memorization vs. Novelty: Training on de-duplicated data (GP-MoLFormer-UNIQ) yields higher novelty (approx. 5-8% higher) than training on raw data, confirming that duplication bias leads to memorization.
- Scaling Law: Novelty decays exponentially with generation size ($y = ae^{-bx}$), yet the model maintains significant novelty (~16.7%) even after generating 10 billion molecules.
Reproducibility Details
Data
- Sources: A combination of PubChem (111M SMILES) and ZINC (1B SMILES) databases.
- Preprocessing:
- All SMILES were canonicalized using RDKit (no isomeric information).
- GP-MoLFormer (Base): Trained on the full 1.1B dataset (includes duplicates).
- GP-MoLFormer-UNIQ: Trained on a de-duplicated subset of 650M SMILES.
- Tokenization: Uses the tokenizer from Schwaller et al. (2019) with a vocabulary size of 2,362 tokens.
- Filtering: Sequences restricted to a maximum length of 202 tokens.
Algorithms
Pair-Tuning (Algorithm 1):
- Objective: Learn task-specific soft prompts $\phi_T$ to maximize $P_{\theta}(b|\phi_T, a)$ where pair $(a, b)$ satisfies property $b > a$.
- Setup: Appends $n$ learnable “enhancement tokens” ($enh$) to the vocabulary.
- Prompt Structure:
<enh_1>...<enh_n><bos><seed_molecule_a><unk><target_molecule_b><eos>. - Training: Freezes base model parameters; updates only enhancement tokens via cross-entropy loss on the target molecule $b$.
- Inference: Prepends learned prompt tokens to seed molecule $a$ and samples autoregressively.
Models
- Architecture: Transformer decoder with Linear Attention (Generalized Random Feature map) and Rotary Positional Embeddings (RoPE).
- Size: ~47M parameters (12 layers, 12 heads, hidden size 768).
- Inference Speed: ~3ms per forward pass on a single A100 GPU.
Evaluation
- Metrics: Validity, Uniqueness, Novelty (MOSES suite), Fréchet ChemNet Distance (FCD), Scaf (Scaffold similarity), SNN (Similarity to Nearest Neighbor).
- Scaling Metrics: Empirical fit for novelty decay: $y = ae^{-bx}$.
Hardware
- Compute: 16 x NVIDIA A100 (80 GB) GPUs across 2 nodes.
- Training Time:
- GP-MoLFormer (1.1B data): ~115 hours total (28.75 hours/epoch for 4 epochs).
- GP-MoLFormer-UNIQ (650M data): ~80 hours total.
- Optimization: Used distributed data-parallel training and adaptive bucketing by sequence length to handle scale.
Paper Information
Citation: Ross, J., Belgodere, B., Hoffman, S. C., Chenthamarakshan, V., Navratil, J., Mroueh, Y., & Das, P. (2025). GP-MoLFormer: A Foundation Model For Molecular Generation. arXiv preprint arXiv:2405.04912. https://doi.org/10.48550/arXiv.2405.04912
Publication: arXiv 2025
@misc{rossGPMoLFormerFoundationModel2025,
title = {{{GP-MoLFormer}}: {{A Foundation Model For Molecular Generation}}},
shorttitle = {{{GP-MoLFormer}}},
author = {Ross, Jerret and Belgodere, Brian and Hoffman, Samuel C. and Chenthamarakshan, Vijil and Navratil, Jiri and Mroueh, Youssef and Das, Payel},
year = 2025,
month = mar,
number = {arXiv:2405.04912},
eprint = {2405.04912},
primaryclass = {q-bio},
publisher = {arXiv},
doi = {10.48550/arXiv.2405.04912},
archiveprefix = {arXiv}
}