What kind of paper is this?
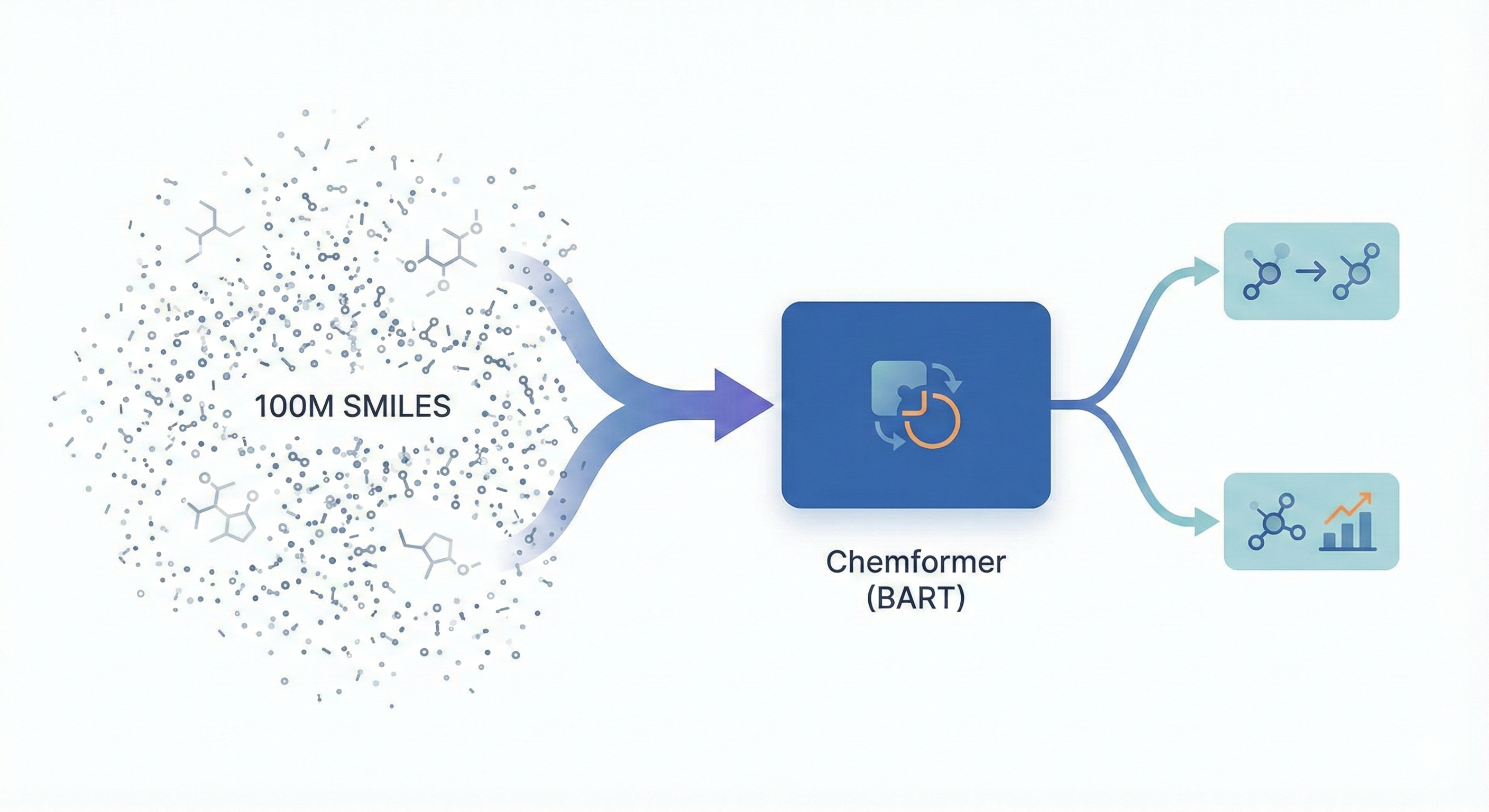
This is a Methodological ($\Psi_{\text{Method}}$) paper. It proposes a novel architecture adaptation (Chemformer based on BART) and a specific pre-training strategy (“Combined” masking and augmentation). The paper validates this method by benchmarking against state-of-the-art models on multiple tasks, including direct synthesis, retrosynthesis, and molecular optimization. It also includes a secondary Resource ($\Psi_{\text{Resource}}$) contribution by making the pre-trained models and code available.
What is the motivation?
Existing Transformer models for cheminformatics are often developed for single applications and are computationally expensive to train from scratch. For example, training a Molecular Transformer for reaction prediction can take days, limiting hyperparameter exploration. While self-supervised pre-training (like BERT or T5) has revolutionized NLP by reducing fine-tuning time and improving performance, its application in chemistry has often been limited to task-specific datasets or encoder-only architectures that struggle with sequence generation. The authors aim to use transfer learning on a large unlabelled dataset to create a model that converges quickly and performs well across diverse sequence-to-sequence and discriminative tasks.
What is the novelty here?
The core novelty lies in the adaptation of the BART architecture for chemistry and the introduction of a “Combined” self-supervised pre-training task.
- Architecture: Chemformer uses the BART encoder-decoder structure, allowing it to handle both discriminative (property prediction) and generative (reaction prediction) tasks efficiently. This provides an alternative to encoder-only (BERT) or decoder-only (GPT) models.
- Combined Pre-training: The authors introduce a task that applies both Span Masking (randomly replacing tokens with
<mask>) and SMILES Augmentation (permuting atom order) simultaneously. - Tunable Augmentation: A novel downstream augmentation strategy is proposed where the probability of augmenting the input/output SMILES ($p_{aug}$) is a tunable hyperparameter, performed on-the-fly.
What experiments were performed?
The authors pre-trained Chemformer on 100 million molecules from ZINC-15 and fine-tuned it on three distinct task types:
- Seq2Seq Reaction Prediction:
- Direct Synthesis: USPTO-MIT dataset (Mixed and Separated).
- Retrosynthesis: USPTO-50K dataset.
- Molecular Optimization: Generating molecules with improved properties (LogD, solubility, clearance) starting from ChEMBL matched molecular pairs.
- Discriminative Tasks:
- QSAR: Predicting properties (ESOL, FreeSolv, Lipophilicity) from MoleculeNet.
- Bioactivity: Predicting pXC50 values for 133 genes using ExCAPE data.
Ablation studies compared three pre-training strategies (Masking, Augmentation, Combined) against a randomly initialized baseline.
What outcomes/conclusions?
- Performance: Chemformer achieved state-of-the-art top-1 accuracy on USPTO-MIT (91.3% Mixed) and USPTO-50K (53.6-54.3%), outperforming the Augmented Transformer and graph-based models (GLN, GraphRetro).
- Convergence Speed: Pre-training significantly accelerated training; fine-tuning for just 20 epochs (30 mins) outperformed the previous SOTA trained for significantly longer.
- Pre-training Tasks: The “Combined” task generally performed best for reaction prediction and bioactivity, while “Masking” was superior for molecular optimization.
- Augmentation Trade-off: The novel augmentation strategy improved top-1 accuracy but degraded top-5/10 accuracy because beam search outputs became populated with augmented versions of the same molecule.
- Discriminative limitations: While pre-training helped, Chemformer did not consistently beat specialized baselines (like D-MPNN or SVR) on small property prediction datasets, suggesting Transformers may require more data or task-specific pre-training to excel here.
Reproducibility Details
Data
The following datasets were used for pre-training and benchmarking.
| Purpose | Dataset | Size | Notes |
|---|---|---|---|
| Pre-training | ZINC-15 | 100M | Selected subset (reactive, purchasable, MW $\le 500$, LogP $\le 5$). Split: 99% Train / 0.5% Val / 0.5% Test. |
| Direct Synthesis | USPTO-MIT | ~470k | Evaluated on “Mixed” and “Separated” variants. |
| Retrosynthesis | USPTO-50K | ~50k | Standard benchmark for retrosynthesis. |
| Optimization | ChEMBL MMPs | ~160k Train | Matched Molecular Pairs for LogD, solubility, and clearance optimization. |
| Properties | MoleculeNet | Small | ESOL (1128), FreeSolv (642), Lipophilicity (4200). |
| Bioactivity | ExCAPE | ~312k | 133 gene targets; >1200 compounds per gene. |
Preprocessing:
- Tokenization: Regex-based tokenization (523 tokens total) derived from ChEMBL 27 canonical SMILES.
- Augmentation: SMILES enumeration (permuting atom order) used for pre-training and on-the-fly during fine-tuning ($p_{aug}=0.5$ for Seq2Seq, $p_{aug}=1.0$ for discriminative).
Algorithms
- Pre-training Tasks:
- Masking: Span masking (BART style).
- Augmentation: Input is a randomized SMILES; target is canonical SMILES.
- Combined: Input is augmented then masked; target is canonical SMILES.
- Optimization:
- Optimizer: Adam ($\beta_1=0.9, \beta_2=0.999$).
- Schedule: Linear warm-up (8000 steps) for pre-training; One-cycle schedule for fine-tuning.
- Inference: Beam search with width 10 for Seq2Seq tasks.
Models
Two model sizes were trained. Both use the Pre-Norm Transformer layout with GELU activation.
| Hyperparameter | Chemformer (Base) | Chemformer-Large |
|---|---|---|
| Layers | 6 | 8 |
| Model Dimension | 512 | 1024 |
| Feed-forward Dim | 2048 | 4096 |
| Attention Heads | 8 | 16 |
| Parameters | ~45M | ~230M |
| Pre-training Task | All 3 variants | Combined only |
Evaluation
Comparisons relied on Top-N accuracy for reaction tasks and validity metrics for optimization.
| Metric | Task | Key Result | Baseline (SOTA) |
|---|---|---|---|
| Top-1 Acc | Direct Synthesis (Sep) | 92.8% (Large) | 91.1% (Aug Transformer) |
| Top-1 Acc | Retrosynthesis | 54.3% (Large) | 53.7% (GraphRetro) / 52.5% (GLN) |
| Desirable % | Mol Optimization | 75.0% (Base-Mask) | 70.2% (Transformer-R) |
| RMSE | Lipophilicity | 0.598 (Combined) | 0.555 (D-MPNN) |
Hardware
- Compute: 4 NVIDIA V100 GPUs (batch size 128 per GPU).
- Training Time:
- Pre-training: 2.5 days (Base) / 6 days (Large) for 1M steps.
- Fine-tuning: ~20-40 epochs for reaction prediction (<12 hours).
Paper Information
Citation: Irwin, R., Dimitriadis, S., He, J., & Bjerrum, E. J. (2022). Chemformer: a pre-trained transformer for computational chemistry. Machine Learning: Science and Technology, 3(1), 015022. https://doi.org/10.1088/2632-2153/ac3ffb
Publication: Machine Learning: Science and Technology 2022
@article{irwinChemformerPretrainedTransformer2022,
title = {Chemformer: A Pre-Trained Transformer for Computational Chemistry},
shorttitle = {Chemformer},
author = {Irwin, Ross and Dimitriadis, Spyridon and He, Jiazhen and Bjerrum, Esben Jannik},
year = 2022,
month = jan,
journal = {Machine Learning: Science and Technology},
volume = {3},
number = {1},
pages = {015022},
publisher = {IOP Publishing},
issn = {2632-2153},
doi = {10.1088/2632-2153/ac3ffb}
}